8UQW
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 13 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8UPQ
 
 | | Campylobacter jejuni ketol-acid reductoisomerase in complex with 2,3-dihydroxy-3-isovalerate. | | Descriptor: | (2R)-2,3-dihydroxy-3-methylbutanoic acid, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Lin, X, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2023-10-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mapping of the Reaction Trajectory catalyzed by Class I Ketol-Acid Reductoisomerase
Acs Catalysis, 2024
|
|
8UQZ
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Gadolinium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GADOLINIUM ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8XJG
 
 | |
6DWV
 
 | | Crystal structure of the LigJ Hydratase in the Apo state | | Descriptor: | 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
8UQX
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8UQY
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Europium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, EUROPIUM (III) ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7JGI
 
 | | NMR structure of the cNTnC-cTnI chimera bound to A7 | | Descriptor: | 7-{[(5-chloronaphthalen-1-yl)sulfonyl]amino}heptanoic acid, CALCIUM ION, Troponin C, ... | | Authors: | Cai, F, Robertson, I.M, Kampourakis, T, Klein, B.A, Sykes, B.D. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Role of Electrostatics in the Mechanism of Cardiac Thin Filament Based Sensitizers.
Acs Chem.Biol., 15, 2020
|
|
6DXQ
 
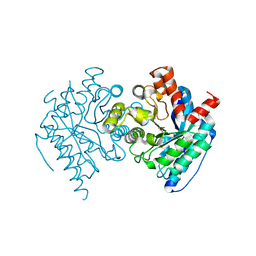 | | Crystal structure of the LigJ Hydratase product complex with 4-carboxy-4-hydroxy-2-oxoadipate | | Descriptor: | (2S)-2-hydroxy-4-oxobutane-1,2,4-tricarboxylic acid, 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
5OLY
 
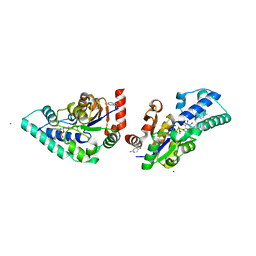 | |
5OLW
 
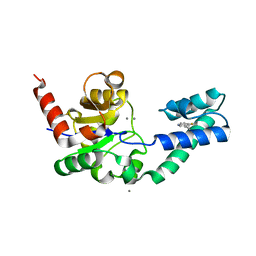 | |
5OLX
 
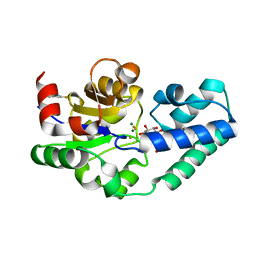 | |
7FJB
 
 | | KpAckA (PduW) with AMPPNP, sodium acetate complex structure | | Descriptor: | ACETATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable propionate kinase, ... | | Authors: | Wenyue, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | KpAckA (PduW) with AMPPNP, sodium acetate complex structure
To Be Published
|
|
7FJ9
 
 | | KpAckA (PduW) with AMPPNP complex structure | | Descriptor: | ACETATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable propionate kinase | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | KpAckA (PduW) with AMPPNP complex structure
To Be Published
|
|
7FJA
 
 | | KpAckA (PduW) with AMPPNP, ethylene glycol complex structure | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable propionate kinase | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | KpAckA (PduW) with AMPPNP, ethylene glycol complex structure
To Be Published
|
|
7JR8
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR6
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JKA
 
 | |
7JIZ
 
 | |
7JI2
 
 | | Crystal Structure of H2-Kb in complex with a OVA mutant peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
7L12
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 14 | | Descriptor: | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L14
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 26 | | Descriptor: | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
