6L3T
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the absence of calcium | | Descriptor: | Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6L12
 
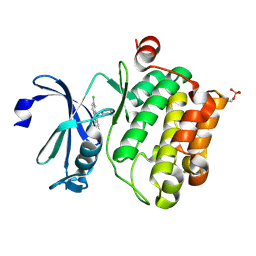 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
7V12
 
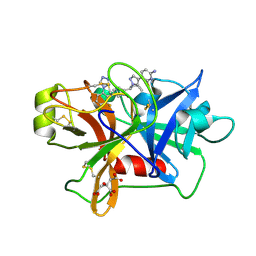 | | Factor XIa in Complex with Compound 2f | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-6-fluoranyl-pyridin-2-amine, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
5V3M
 
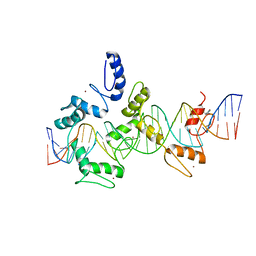 | | mouseZFP568-ZnF1-11 in complex with DNA | | Descriptor: | DNA (28-MER), ZINC ION, Zinc finger protein 568 | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
7V0Z
 
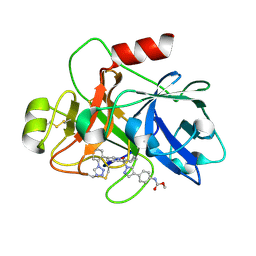 | | Factor XIa in Complex with Compound 2a | | Descriptor: | Coagulation factor XIa light chain, methyl ~{N}-[4-[1-[(1~{R})-1-[5-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]phenyl]carbamate | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
6L3A
 
 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
7MC3
 
 | | Solution structure of Miz-1 zinc finger 12 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
7V16
 
 | | Factor XIa in Complex with Compound 2j | | Descriptor: | 5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-2-[(1~{R})-2-cyclopropyl-1-[4-(2-methylpyrazol-3-yl)pyrazol-1-yl]ethyl]-1-oxidanyl-pyridine, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
7DD0
 
 | |
6BP9
 
 | |
7MC1
 
 | | Solution structure of Miz-1 Zinc finger 10 | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
5V80
 
 | |
7V15
 
 | | Factor XIa in Complex with Compound 2i | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-4-methyl-1,3-thiazole, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Cedervall, P, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
7V11
 
 | | Factor XIa in Complex with Compound 2e | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-6-methyl-pyridin-2-amine, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
6L9O
 
 | | Crystal structure of FABP7 apo | | Descriptor: | Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of FABP7 apo
To Be Published
|
|
5UND
 
 | |
7V10
 
 | | Factor XIa in Complex with Compound 2d | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, methyl ~{N}-[4-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]phenyl]carbamate | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
5V8V
 
 | | Crystal Structure of Human Renin in Complex with a biphenylpipderidinylcarbinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, methyl [(4S)-4-(3'-ethyl-6-fluoro[1,1'-biphenyl]-2-yl)-4-hydroxy-4-{(3R)-1-[4-(methylamino)butanoyl]piperidin-3-yl}butyl]carbamate | | Authors: | Concha, N, Zhao, B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of renin inhibitors containing a simple aspartate binding moiety that imparts reduced P450 inhibition.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7MC2
 
 | | Solution structure of Miz-1 Zinc finger 11 H586Y | | Descriptor: | Isoform 2 of Zinc finger and BTB domain-containing protein 17, ZINC ION | | Authors: | Boisvert, O, Lavigne, P. | | Deposit date: | 2021-04-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc Fingers 10 and 11 of Miz-1 undergo conformational exchange to achieve specific DNA binding.
Structure, 30, 2022
|
|
8GSF
 
 | | Echovirus3 empty particle in complex with 6D10 Fab (sideling) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
6L8I
 
 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LAU
 
 | | the wildtype SAM-VI riboswitch bound to SAH | | Descriptor: | CESIUM ION, GUANOSINE-5'-TRIPHOSPHATE, RNA (54-MER), ... | | Authors: | Ren, A, Sun, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
8GSE
 
 | | Echovirus3 capsid protein in complex with 6D10 Fab (upright) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
5VC0
 
 | | Crystal structure of human CYP3A4 bound to ritonavir | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, RITONAVIR | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-03-30 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | High-Level Production and Properties of the Cysteine-Depleted Cytochrome P450 3A4.
Biochemistry, 56, 2017
|
|
