7EOK
 
 | | Crystal structure of the Pepper aptamer in complex with HBC485 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-(dimethylamino)ethyl-methyl-amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOO
 
 | | Crystal structure of the Pepper aptamer in complex with HBC525 | | Descriptor: | (~{E})-2-(1,3-benzoxazol-2-yl)-3-[4-[2-hydroxyethyl(methyl)amino]phenyl]prop-2-enenitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOI
 
 | |
7EOP
 
 | | Crystal structure of the Pepper aptamer in complex with HBC620 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EON
 
 | | Crystal structure of the Pepper aptamer in complex with HBC514 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-(dimethylamino)ethyl-methyl-amino]phenyl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOG
 
 | |
7EOL
 
 | | Crystal structure of the Pepper aptamer in complex with HBC497 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOJ
 
 | |
7EOM
 
 | | Crystal structure of the Pepper aptamer in complex with HBC508 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[6-[2-hydroxyethyl(methyl)amino]pyridin-3-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOH
 
 | | Crystal structure of the Pepper aptamer in complex with HBC | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-hydroxyethyl(methyl)amino]phenyl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, Pepper (49-MER) | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
5OL9
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) N-terminal domain | | Descriptor: | ACETATE ION, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5OLA
 
 | | Structure of mitochondrial transcription elongation complex in complex with elongation factor TEFM | | Descriptor: | DNA (30-MER), DNA (5'-D(P*AP*TP*GP*GP*TP*GP*TP*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*GP*AP*AP*C)-3'), DNA-directed RNA polymerase, ... | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.904 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5OL8
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) C-terminal domain | | Descriptor: | GLYCEROL, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
8AYG
 
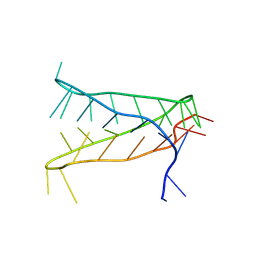 | | Crystal structure of an intramolecular i-motif at the insulin-linked polymorphic region (ILPR) | | Descriptor: | Insulin-linked polymorphic region, ILPR DNA (31-MER) | | Authors: | Parkinson, G.N, Alexandrou, E, Waller, Z.A.E, El-Omari, K. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into i-motif DNA structures in sequences from the insulin-linked polymorphic region.
Nat Commun, 15, 2024
|
|
4P1D
 
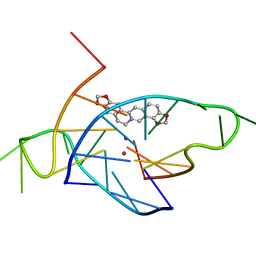 | | Structure of the complex of a bimolecular human telomeric DNA with Coptisine | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, POTASSIUM ION, telomeric DNA | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the complex of a bimolecular human telomeric DNA with Coptisine
to be published
|
|
6Z8X
 
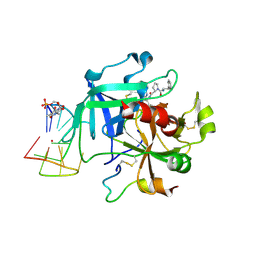 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6Z8V
 
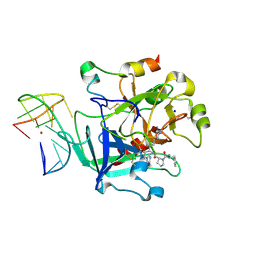 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
7U0Y
 
 | |
2RNK
 
 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
7SZU
 
 | |
7OA3
 
 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium hexammine co-crystallized form) | | Descriptor: | Chili RNA Aptamer, DMHBO+, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAX
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBO+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAW
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBI+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBI+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAV
 
 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium III hexammine soaking crystal form) | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
6Z8W
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3G), which contains 1-beta-D-glucopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
