8XMB
 
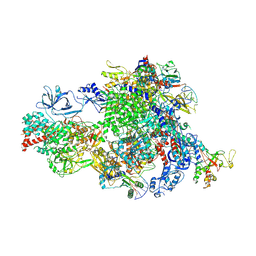 | | NTP-bound Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XMD
 
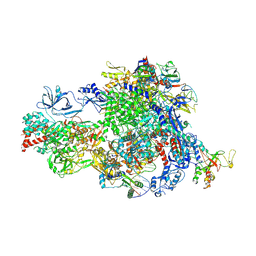 | | Pre-translocated Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XME
 
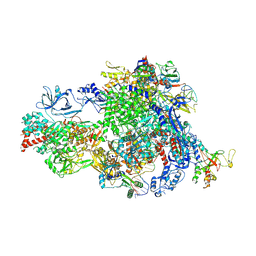 | | Backtracked Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
5EZK
 
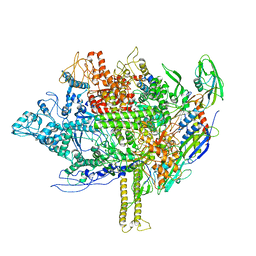 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
2NVT
 
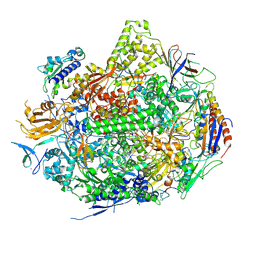 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with GMPCPP | | Descriptor: | 5'-D(*GP*TP*AP*CP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*TP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2F8E
 
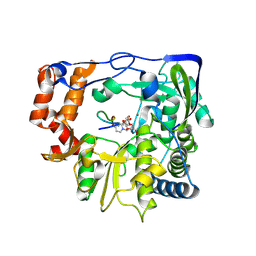 | | Foot and Mouth Disease Virus RNA-dependent RNA polymerase in complex with uridylylated VPg protein | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA-dependent RNA polymerase, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2005-12-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a protein primer-polymerase complex in the initiation of genome replication
Embo J., 25, 2006
|
|
7PYK
 
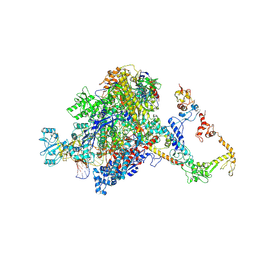 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY7
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in more-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY5
 
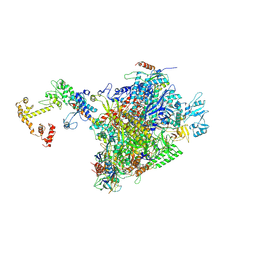 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (the consensus NusA-NusG-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY3
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (the consensus NusA-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PYJ
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA (NusA elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-10 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY6
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (NusA and NusG elongation complex in less-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1HHS
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
3P9Y
 
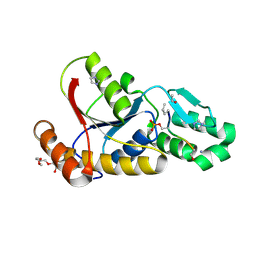 | |
3TJ1
 
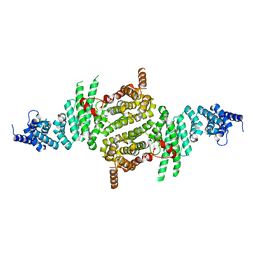 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
7NVX
 
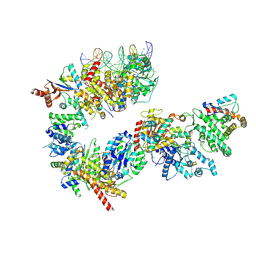 | | TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVW
 
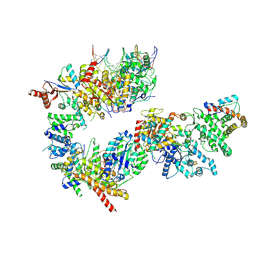 | | TFIIH in a pre-translocated state (without ADP-BeF3) | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription factor IIE subunit 1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
3JB7
 
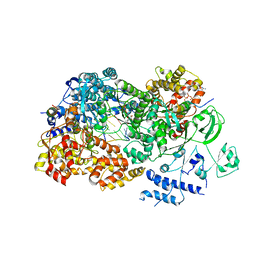 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
7NVV
 
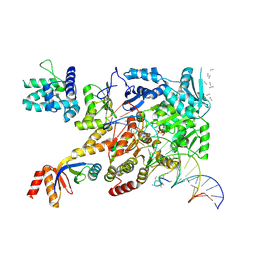 | | XPB-containing part of TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
8A8F
 
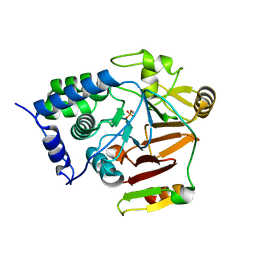 | | Crystal structure of Glc7 phosphatase in complex with the regulatory region of Ref2 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, RNA end formation protein 2, ... | | Authors: | Carminati, M, Manav, C.M, Bellini, D, Passmore, L.A. | | Deposit date: | 2022-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription.
Mol.Cell, 83, 2023
|
|
7ZSA
 
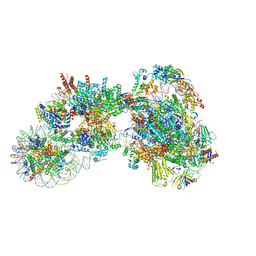 | |
7ZS9
 
 | |
7ZSB
 
 | |
8K5P
 
 | |
