1D6O
 
 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1CI7
 
 | | TERNARY COMPLEX OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Anderson, A.C, O'Neil, R.H, Delano, W.L, Stroud, R.M. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural mechanism for half-the-sites reactivity in an enzyme, thymidylate synthase, involves a relay of changes between subunits.
Biochemistry, 38, 1999
|
|
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2NV0
 
 | |
2LDC
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP1 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2OH6
 
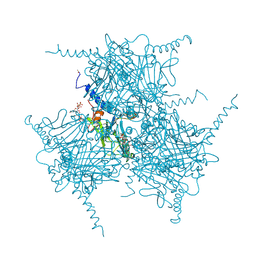 | | The Crystal Structure of Recombinant Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Coulibaly, F, Chiu, E, Ikeda, K, Gutmann, S, Haebel, P.W, Schulze-Briese, C, Mori, H, Metcalf, P. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular organization of cypovirus polyhedra.
Nature, 446, 2007
|
|
2NQZ
 
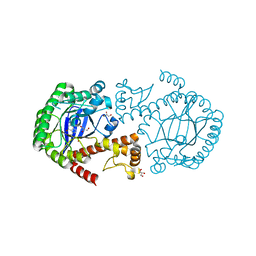 | | Trna-guanine transglycosylase (TGT) mutant in complex with 7-deaza-7-aminomethyl-guanine | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
2M89
 
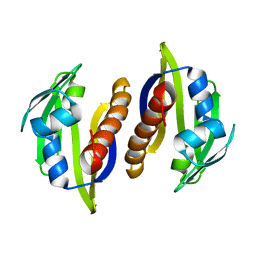 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2LHD
 
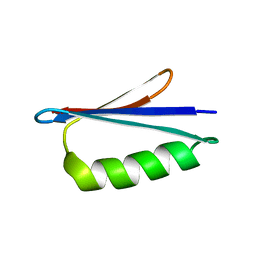 | | GB98 solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LDA
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2NV1
 
 | | Structure of the synthase subunit Pdx1 (YaaD) of PLP synthase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Strohmeier, M, Tews, I, Sinning, I. | | Deposit date: | 2006-11-10 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of a bacterial pyridoxal 5'-phosphate synthase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2OH5
 
 | | The Crystal Structure of Infectious Cypovirus Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Coulibaly, F, Chiu, E, Ikeda, K, Gutmann, S, Haebel, P.W, Schulze-Briese, C, Mori, H, Metcalf, P. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The molecular organization of cypovirus polyhedra.
Nature, 446, 2007
|
|
3TJF
 
 | | Crystal Structure of human peroxiredoxin IV C51A mutant in reduced form | | Descriptor: | Peroxiredoxin-4, SULFATE ION | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
3STU
 
 | |
3T90
 
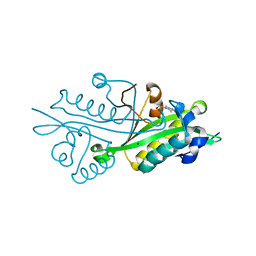 | | Crystal structure of glucosamine-6-phosphate N-acetyltransferase from Arabidopsis thaliana | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glucose-6-phosphate acetyltransferase 1, SODIUM ION | | Authors: | Grishkovskaya, I, Herter, T, Riegler, H, Usadel, B. | | Deposit date: | 2011-08-02 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and functional characterization of a glucosamine-6-phosphate N-acetyltransferase from Arabidopsis thaliana.
Biochem.J., 443, 2012
|
|
3T58
 
 | |
6J3Z
 
 | | Structure of C2S1M1-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
3STY
 
 | |
6J3Y
 
 | | Structure of C2S2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-01-07 | | Release date: | 2019-08-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex.
Nat.Plants, 5, 2019
|
|
3T59
 
 | |
6JLP
 
 | | XFEL structure of cyanobacterial photosystem II (3F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLN
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
3USR
 
 | | Structure of Y194F glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Issoglio, F.M, Carrizo, M.E, Romero, J.M, Curtino, J.A. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of monomeric and dimeric glycogenin autoglucosylation.
J.Biol.Chem., 287, 2012
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | Descriptor: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
