3C9Q
 
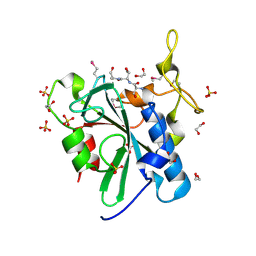 | | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the uncharacterized human protein C8orf32 with bound peptide.
To be Published
|
|
3CAS
 
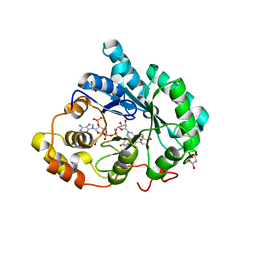 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 4-androstenedione | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, 4-ANDROSTENE-3-17-DIONE, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
3CBG
 
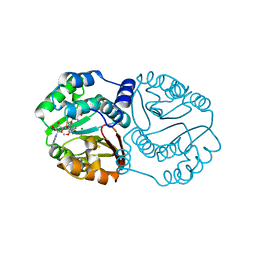 | | Functional and Structural Characterization of a Cationdependent O-Methyltransferase from the Cyanobacterium Synechocystis Sp. Strain PCC 6803 | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)prop-2-enoic acid, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, ... | | Authors: | Kopycki, J.G, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-02-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Characterization of a Cation-dependent O-Methyltransferase from the Cyanobacterium Synechocystis sp. Strain PCC 6803
J.Biol.Chem., 283, 2008
|
|
3CBQ
 
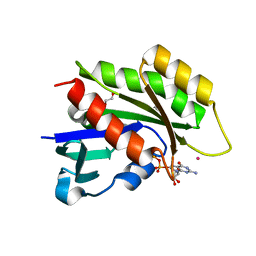 | | Crystal structure of the human REM2 GTPase with bound GDP | | Descriptor: | 1,2-ETHANEDIOL, GTP-binding protein REM 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nedyalkova, L, Shen, Y, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the human REM2 GTPase with bound GDP.
To be Published
|
|
3ATD
 
 | |
3AUO
 
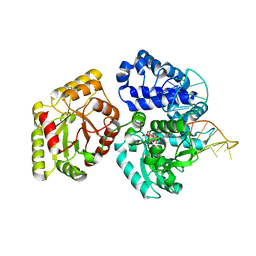 | | DNA polymerase X from Thermus thermophilus HB8 ternary complex with 1-nt gapped DNA and ddGTP | | Descriptor: | 1-nt gapped DNA, 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase beta family (X family), ... | | Authors: | Nakane, S, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA.
J.Mol.Biol., 417, 2012
|
|
3AS5
 
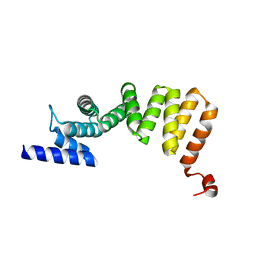 | | MamA AMB-1 P212121 | | Descriptor: | MAGNESIUM ION, MamA | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-10 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AST
 
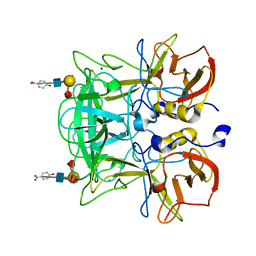 | | Crystal structure of P domain Q389N mutant from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASV
 
 | | The Closed form of serine dehydrogenase complexed with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHATE ION, Short-chain dehydrogenase/reductase SDR | | Authors: | Yamazawa, R, Nakajima, Y, Yoshimoto, T, Ito, K. | | Deposit date: | 2010-12-21 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of serine dehydrogenase from Escherichia coli: important role of the C-terminal region for closed-complex formation.
J.Biochem., 149, 2011
|
|
3ATS
 
 | | Crystal structure of Rv3168 | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y.-G, Kim, S, Nguyen, C.M.T, Kim, K.-J. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv3168: a putative aminoglycoside antibiotics resistance enzyme
Proteins, 79, 2011
|
|
3ASU
 
 | |
3ATE
 
 | |
3RLC
 
 | |
3AW5
 
 | | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3AWM
 
 | | Cytochrome P450SP alpha (CYP152B1) wild-type with palmitic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position
J.Biol.Chem., 286, 2011
|
|
3AZ1
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | Vitamin D3 receptor, {4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetic acid | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AWP
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant F288G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
3AZ2
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | 5-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ3
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[4-(3-{4-[(3S)-3-hydroxy-4,4-dimethylpentyl]-3-methylphenyl}pentan-3-yl)-2-methylphenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AWQ
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant L78F | | Descriptor: | Fatty acid alpha-hydroxylase, PALMITIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
3CC0
 
 | |
3CBB
 
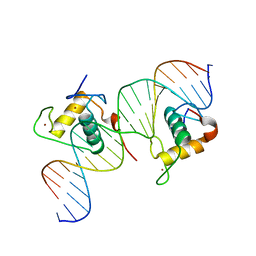 | | Crystal Structure of Hepatocyte Nuclear Factor 4alpha in complex with DNA: Diabetes Gene Product | | Descriptor: | Hepatocyte Nuclear Factor 4-alpha promoter element DNA, Hepatocyte Nuclear Factor 4-alpha, DNA binding domain, ... | | Authors: | Lu, P, Rha, G.B, Melikishvili, M, Adkins, B.C, Fried, M.G, Chi, Y.I. | | Deposit date: | 2008-02-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of natural promoter recognition by a unique nuclear receptor, HNF4alpha. Diabetes gene product.
J.Biol.Chem., 283, 2008
|
|
3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
3RBL
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | CHLORIDE ION, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RKO
 
 | |
