1S5Q
 
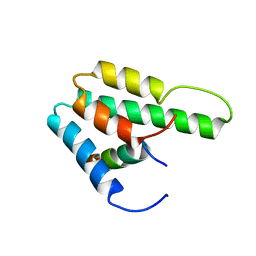 | | Solution Structure of Mad1 SID-mSin3A PAH2 Complex | | Descriptor: | MAD protein, Sin3a protein | | Authors: | Swanson, K.A, Knoepfler, P.S, Huang, K, Kang, R.S, Cowley, S.M, Laherty, C.D, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2004-01-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2JS7
 
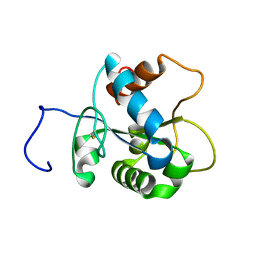 | | Solution NMR structure of human myeloid differentiation primary response (MyD88). Northeast Structural Genomics target HR2869A | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Rossi, P, Ramelot, T.A, Tao, X, Ciano, M, Ho, C, Ma, L.-C, Xiao, R, Acton, T.B, Kennedy, M.A, Tong, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Myeloid Differentiation Primary Response (MyD88).
To be Published
|
|
1S7L
 
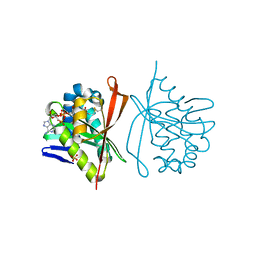 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase in complex with Coenzyme A (CoA-Cys134 Disulfide) | | Descriptor: | COENZYME A, SULFATE ION, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
1LCZ
 
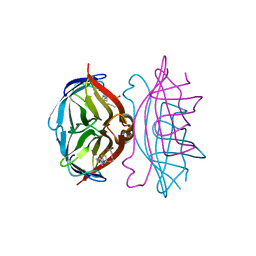 | | streptavidin-BCAP complex | | Descriptor: | E-AMINO BIOTINYL CAPROIC ACID, Streptavidin | | Authors: | Livnah, O, Pazy, Y, Bayer, E.A, Wilchek, M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1LDQ
 
 | | avidin-homobiotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, HOMOBIOTIN | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-09 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
3V16
 
 | | An intramolecular pi-cation latch in phosphatidylinositol-specific phospholipase C from S.aureus controls substrate access to the active site | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
1S7N
 
 | | Ribosomal L7/L12 alpha-N-protein acetyltransferase in complex with Coenzyme A (CoA free sulfhydryl) | | Descriptor: | COENZYME A, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
1S7K
 
 | |
2JQ9
 
 | | VPS4A MIT-CHMP1A complex | | Descriptor: | Chromatin-modifying protein 1a, Vacuolar protein sorting-associating protein 4A | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JQK
 
 | | VPS4B MIT-CHMP2B Complex | | Descriptor: | Charged multivesicular body protein 2b, Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JXP
 
 | | Solution NMR structure of uncharacterized lipoprotein B from Nitrosomonas europaea. Northeast Structural Genomics target NeR45A | | Descriptor: | Putative lipoprotein B | | Authors: | Rossi, P, Wang, D, Janjua, H, Owens, L, Xiao, R, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of uncharacterized lipoprotein B from Nitrosomonas europaea.
To be Published
|
|
1RXH
 
 | | Crystal structure of streptavidin mutant L124R (M1) complexed with biotinyl p-nitroanilide (BNI) | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
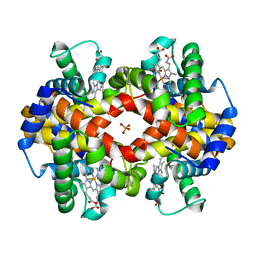
1MKO
 
 | |
6Z9E
 
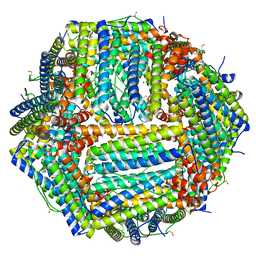 | | 1.55 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6Z6F
 
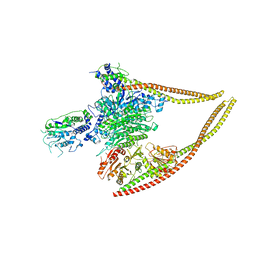 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
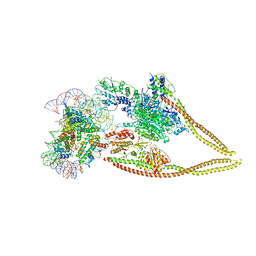 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z71
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZSI
 
 | | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members. | | Descriptor: | DI(HYDROXYETHYL)ETHER, EH domain-binding protein 1, MAGNESIUM ION, ... | | Authors: | Rai, A, Bleimling, N, Vetter, I.R, Goody, R.S. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members.
Nat Commun, 11, 2020
|
|
6ZSJ
 
 | | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members. | | Descriptor: | EH domain-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Rai, A, Bleimling, N, Vetter, I.R, Goody, R.S. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of activation of the actin binding protein EHBP1 by Rab8 family members.
Nat Commun, 11, 2020
|
|
7A6A
 
 | | 1.15 A structure of human apoferritin obtained from Titan Mono- BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Chari, A, Stark, H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-09-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.15 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
7A8W
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with an engineered HMA domain of Pikp-1 from rice (Oryza sativa) | | Descriptor: | NBS-LRR class disease resistance protein, Uncharacterized protein | | Authors: | Maidment, J.H.R, De la Concepcion, J.C, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
1WRI
 
 | | Crystal Structure of Ferredoxin isoform II from E. arvense | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin II | | Authors: | Kurisu, G, Nishiyama, D, Kusunoki, M, Fujikawa, S, Katoh, M, Hanke, G.T, Hase, T, Teshima, K. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A structural basis of Equisetum arvense ferredoxin isoform II producing an alternative electron transfer with ferredoxin-NADP+ reductase.
J.Biol.Chem., 280, 2005
|
|
