1E8C
 
 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1E8D
 
 | | MECHANISTIC ASPECTS OF CYANOGENESIS FROM ACTIVE SITE MUTANT SER80ALA OF HYDROXYNITRILE LYASE FROM MANIHOT ESCULENTA IN COMPLEX WITH ACETONE CYANOHYDRIN | | Descriptor: | 2-HYDROXY-2-METHYLPROPANENITRILE, HYDROXYNITRILE LYASE | | Authors: | Lauble, H, Miehlich, B, Foerster, S, Wajant, H, Effenberger, F. | | Deposit date: | 2000-09-19 | | Release date: | 2001-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Aspects of Cyanogenesis from Active-Site Mutant Ser80Ala of Hydroxynitrile Lyase from Manihot Esculenta in Complex with Acetone Cyanohydrin.
Protein Sci., 10, 2001
|
|
1E8E
 
 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
1E8F
 
 | |
1E8G
 
 | |
1E8H
 
 | |
1E8I
 
 | | HUMAN CD69 - TETRAGONAL FORM | | Descriptor: | EARLY ACTIVATION ANTIGEN CD69, SULFATE ION | | Authors: | Tormo, J. | | Deposit date: | 2000-09-21 | | Release date: | 2000-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the C-Type Lectin-Like Domain from the Human Hematopoietic Cell Receptor Cd69
J.Biol.Chem., 276, 2001
|
|
1E8J
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
1E8K
 
 | | Cyclophilin 3 Complexed With Dipeptide Ala-Pro | | Descriptor: | ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 3, PROLINE | | Authors: | Wu, S.Y, Dornan, J, Kontopidis, G, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Direct Determination of a Ligand Binding Constant in Protein Crystals
Angew.Chem.Int.Ed.Engl., 40, 2001
|
|
1E8L
 
 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
1E8M
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT, COMPLEXED WITH INHIBITOR | | Descriptor: | GLYCEROL, N-[(benzyloxy)carbonyl]glycyl-L-proline, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-09-27 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Prolyl Oligopeptidase Substrate/ Inhibitor Complexes. Use of Inhibitor Binding for Titration of the Catalytic Histidine Residue
J.Biol.Chem., 276, 2001
|
|
1E8N
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT, COMPLEXED WITH PEPTIDE | | Descriptor: | GLYCEROL, PEPTIDE INHIBITOR, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-09-27 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Prolyl Oligopeptidase Substrate/ Inhibitor Complexes. Use of Inhibitor Binding for Titration of the Catalytic Histidine Residue
J.Biol.Chem., 276, 2001
|
|
1E8O
 
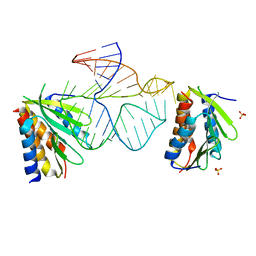 | | Core of the Alu domain of the mammalian SRP | | Descriptor: | 7SL RNA, SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Weichenrieder, O, Wild, K, Strub, K, Cusack, S. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Assembly of the Alu Domain of the Mammalian Signal Recognition Particle
Nature, 408, 2000
|
|
1E8P
 
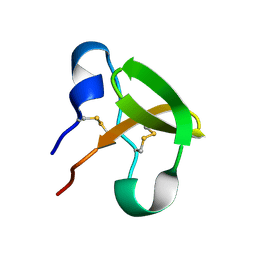 | | Characterisation of the cellulose docking domain from Piromyces equi | | Descriptor: | Endoglucanase 45A | | Authors: | Raghothama, S, Eberhardt, R.Y, White, P, Hazlewood, G.P, Gilbert, H.J, Simpson, P.J, Williamson, M.P. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-07 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi.
Nat. Struct. Biol., 8, 2001
|
|
1E8Q
 
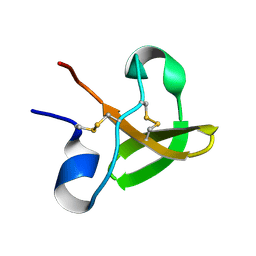 | | Characterisation of the cellulose docking domain from Piromyces equi | | Descriptor: | Endoglucanase 45A | | Authors: | Raghothama, S, Eberhardt, R.Y, White, P, Hazlewood, G.P, Gilbert, H.J, Simpson, P.J, Williamson, M.P. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-07 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi.
Nat. Struct. Biol., 8, 2001
|
|
1E8R
 
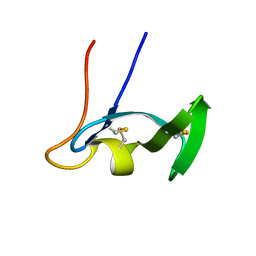 | | SOLUTION STRUCTURE OF TYPE X CBD | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Raghothama, S, Simpson, P.J, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-09-28 | | Release date: | 2000-10-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cbm10 Cellulose Binding Module from Pseudomonas Xylanase A
Biochemistry, 39, 2000
|
|
1E8S
 
 | | Alu domain of the mammalian SRP (potential Alu retroposition intermediate) | | Descriptor: | 7SL RNA, 88-MER, EUROPIUM (III) ION, ... | | Authors: | Weichenrieder, O, Wild, K, Strub, K, Cusack, S. | | Deposit date: | 2000-09-29 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Assembly of the Alu Domain of the Mammalian Signal Recognition Particle
Nature, 408, 2000
|
|
1E8T
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8U
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8V
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8W
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E8X
 
 | | STRUCTURAL INSIGHTS INTO PHOSHOINOSITIDE 3-KINASE ENZYMATIC MECHANISM AND SIGNALLING | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LUTETIUM (III) ION, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E8Y
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E8Z
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT, STAUROSPORINE | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E90
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
