4YA6
 
 | | Crystal structure of LigO-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
7FQT
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRP
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with XST00000245b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(2-methyl-1,3-thiazol-4-yl)thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
5U96
 
 | |
7RXL
 
 | |
7FQM
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000619a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-({[(thiophen-2-yl)methyl]amino}methyl)phenol, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
4NZZ
 
 | |
1K0F
 
 | | Crystal structure of Zn(II)-free T. pallidum TroA | | Descriptor: | Periplasmic zinc-binding protein troA | | Authors: | Lee, Y.H, Dorwart, M.R, Hazlett, K.R, Deka, R.K, Norgard, M.V, Radolf, J.D, Hasemann, C.A. | | Deposit date: | 2001-09-19 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation.
J.Bacteriol., 184, 2002
|
|
8R7Z
 
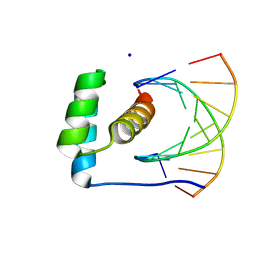 | | transcription factor BARHL2 homodimer with spacing four bp | | Descriptor: | BarH-like 2 homeobox protein, DNA (5'-D(P*AP*CP*CP*GP*TP*TP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*AP*AP*CP*GP*GP*T)-3'), ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 2025
|
|
4O43
 
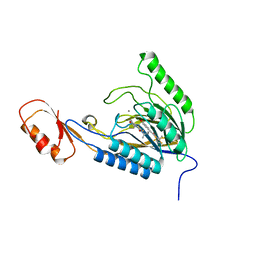 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-3-[(2~{R})-butan-2-yl]-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
2WX0
 
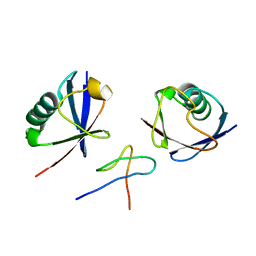 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked di-ubiquitin, P21 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
7FQV
 
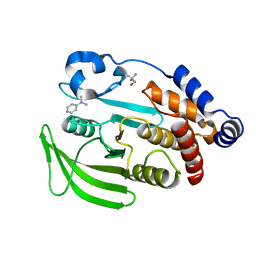 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with XST00000847b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N,N-dimethylpyridin-4-amine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
6FJ5
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-AGG-HG) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2018-01-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
7FQX
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000601a | | Descriptor: | 1-(3,4-dimethoxyphenyl)methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FQU
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMSOA000470b | | Descriptor: | (3-chlorophenoxy)acetic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
4BOQ
 
 | | Structure of OTUD2 OTU domain | | Descriptor: | GLYCEROL, UBIQUITIN THIOESTERASE OTU1 | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
8T0W
 
 | | Crystal structure of dimethylsulfone (DMSO2) monooxygenase SfnG from Pseudomonas fluorescens with DMSO2 and oxidized FMN bound | | Descriptor: | (methanesulfonyl)methane, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gonzalez, R, Soule, J, Dowling, D.P. | | Deposit date: | 2023-06-01 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, biophysical, and biochemical insights into C-S bond cleavage by dimethylsulfone monooxygenase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7FRF
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000089a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, methyl 3-(methylsulfonylamino)benzoate | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRH
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with Z2856434762 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N,N-dimethylpyridin-4-amine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
5HAF
 
 | |
4DTK
 
 | | Novel and selective pan-PIM kinase inhibitor | | Descriptor: | (5Z)-5-{2-[(3R)-3-aminopiperidin-1-yl]-3-(propan-2-yloxy)benzylidene}-1,3-thiazolidine-2,4-dione, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2012-02-21 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of novel benzylidene-1,3-thiazolidine-2,4-diones as potent and selective inhibitors of the PIM-1, PIM-2, and PIM-3 protein kinases.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7FQS
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000555a | | Descriptor: | (2R,5R,6S)-2,3,4,5,6,7-hexahydro-1H-2,6-methanoazocino[5,4-b]indol-5-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FQY
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000278a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}1-(4,6-dimethylpyrimidin-2-yl)benzene-1,4-diamine | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRL
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with Z2856434917 | | Descriptor: | 1-(3,4-dimethoxyphenyl)methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FQO
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000523a | | Descriptor: | (1R,4R,5R,6S)-2-(methanesulfonyl)-4,6-dimethoxy-2-azabicyclo[3.3.1]nonane, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
