6ZHA
 
 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4R63
 
 | | Binary complex crystal structure of R258A mutant of DNA polymerase Beta | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
4R64
 
 | | Binary complex crystal structure of E295K mutant of DNA polymerase Beta | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
2V6Z
 
 | | Solution Structure of Amino-Terminal Domain of Human DNA Polymerase Epsilon Subunit B | | Descriptor: | DNA POLYMERASE EPSILON SUBUNIT 2 | | Authors: | Nuutinen, T, Fredriksson, K, Tossavainen, H, Pospiech, H, Pirila, P, Permi, P, Annila, A, Syvaoja, J.E. | | Deposit date: | 2007-07-24 | | Release date: | 2008-08-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Amino-Terminal Domain of Human DNA Polymerase Epsilon Subunit B is Homologous to C-Domains of Aaa+ Proteins.
Nucleic Acids Res., 36, 2008
|
|
5V1G
 
 | | DNA polymerase beta binary complex with 8-oxoG at the primer terminus | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Smith, M.R, Whitaker, A.M, Schaich, M.A. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
5V1H
 
 | | DNA polymerase beta binary complex with 8-oxoG:A at the primer terminus | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Schaich, M.A, Whitaker, A.M, Smith, M.R. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
7Z87
 
 | | DNA-PK in the active state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z88
 
 | | DNA-PK in the intermediate state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5V1J
 
 | | DNA polymerase beta open product complex with 8-oxoG:C and inserted dCTP | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG)P*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Smith, M.R, Whitaker, A.M, Schaich, M.A. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
6BTE
 
 | | DNA Polymerase Beta I260Q Binary Complex | | Descriptor: | CHLORIDE ION, DNA Downstream Strand, DNA Primer Strand, ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2017-12-06 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | I260Q DNA polymerase beta highlights precatalytic conformational rearrangements critical for fidelity.
Nucleic Acids Res., 46, 2018
|
|
5U8I
 
 | | DNA Polymerase Beta S229L crystallized in PEG 400 | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Remote Mutations Induce Functional Changes in Active Site Residues of Human DNA Polymerase beta.
Biochemistry, 56, 2017
|
|
6UHN
 
 | | Crystal Structure of C148 mGFP-cDNA-1 | | Descriptor: | C148 mGFP-cDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHQ
 
 | | Crystal Structure of C148 mGFP-cDNA-3 | | Descriptor: | C148 mGFP-cDNA-3, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHR
 
 | | Crystal Structure of C148 mGFP-scDNA-2 | | Descriptor: | C148 mGFP-scDNA-2 | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHP
 
 | | Crystal Structure of C148 mGFP-ncDNA-1 | | Descriptor: | C148 mGFP-ncDNA-1 | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHL
 
 | | Crystal Structure of C148 mGFP-scDNA-1 | | Descriptor: | C148 mGFP-scDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6UHO
 
 | | Crystal Structure of C148 mGFP-cDNA-2 | | Descriptor: | C148 mGFP-cDNA-2, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6BTF
 
 | | DNA Polymerase Beta I260Q Ternary Complex | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA Downstream Strand, DNA Primer Strand, ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2017-12-06 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | I260Q DNA polymerase beta highlights precatalytic conformational rearrangements critical for fidelity.
Nucleic Acids Res., 46, 2018
|
|
5UGP
 
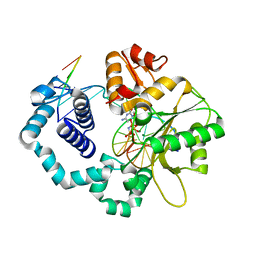 | | DNA polymerase beta complex with a 1nt gap and dCMPPNP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]cytidine, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2017-01-09 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Modulating the DNA polymerase beta reaction equilibrium to dissect the reverse reaction.
Nat. Chem. Biol., 13, 2017
|
|
5UGN
 
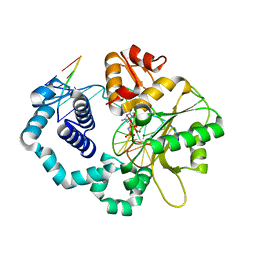 | | DNA polymerase beta imidodiphosphate reactant complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]cytidine, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2017-01-09 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Modulating the DNA polymerase beta reaction equilibrium to dissect the reverse reaction.
Nat. Chem. Biol., 13, 2017
|
|
3VDY
 
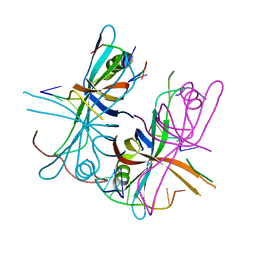 | | B. subtilis SsbB/ssDNA | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein ssbB | | Authors: | Yadav, T, Carrasco, B, Myers, A.R, George, N.P, Keck, J.L, Alonso, J.C. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins.
Nucleic Acids Res., 40, 2012
|
|
8QQS
 
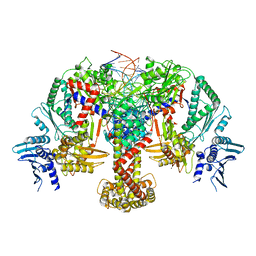 | |
8QQU
 
 | |
1QZH
 
 | | Crystal structure of Pot1 (protection of telomere)- ssDNA complex | | Descriptor: | Protection of telomeres protein 1, telomeric single-stranded DNA | | Authors: | Lei, M, Podell, E.R, Baumann, P, Cech, T.R. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA
Nature, 426, 2003
|
|
8QDX
 
 | | E. coli DNA gyrase bound to a DNA crossover | | Descriptor: | DNA gyrase subunit A, DNA gyrase subunit B, DNA minicircle | | Authors: | Vayssieres, M, Lamour, V. | | Deposit date: | 2023-08-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of DNA crossover capture by Escherichia coli DNA gyrase.
Science, 384, 2024
|
|
