7JMA
 
 | |
1RW4
 
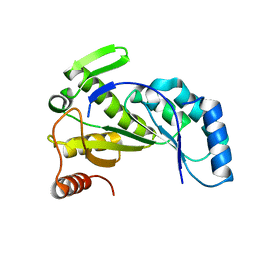
 | | Nitrogenase Fe protein l127 deletion variant | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Sen, S, Igarashi, R, Smith, A, Johnson, M.K, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Mimic of the MgATP-Bound "On State" of the Nitrogenase Iron Protein.
Biochemistry, 43, 2004
|
|
1M34
 
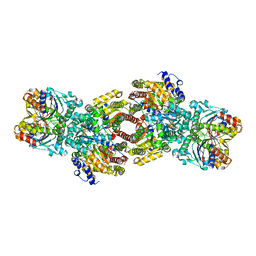 | | Nitrogenase Complex From Azotobacter Vinelandii Stabilized By ADP-Tetrafluoroaluminate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.-J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4 Stabilized structure
Biochemistry, 41, 2002
|
|
1N2C
 
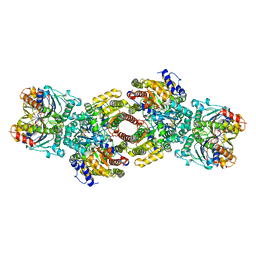 | | NITROGENASE COMPLEX FROM AZOTOBACTER VINELANDII STABILIZED BY ADP-TETRAFLUOROALUMINATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schindelin, H, Kisker, C, Rees, D.C. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction.
Nature, 387, 1997
|
|
8ENO
 
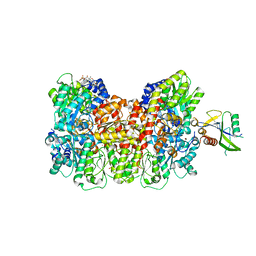 | | Homocitrate-deficient nitrogenase MoFe-protein from A. vinelandii nifV knockout in complex with NafT | | Descriptor: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8ENM
 
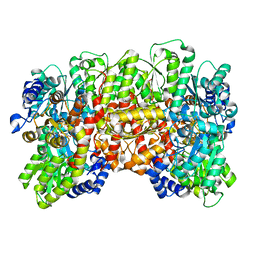 | | CryoEM structure of the high pH nitrogenase MoFe-protein under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8Q50
 
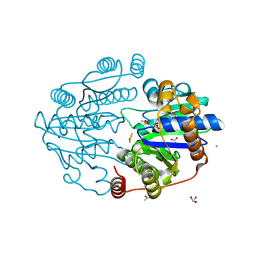 | |
8ENL
 
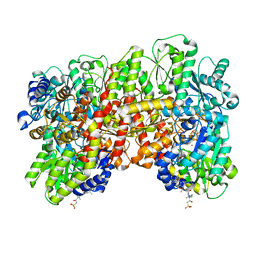 | | CryoEM structure of the high pH turnover-inactivated nitrogenase MoFe-protein | | Descriptor: | CHAPSO, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8ENN
 
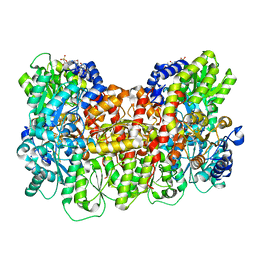 | | Homocitrate-deficient nitrogenase MoFe-protein from Azotobacter vinelandii nifV knockout | | Descriptor: | CHAPSO, CITRIC ACID, FE (III) ION, ... | | Authors: | Warmack, R.A, Maggiolo, A.O, Rees, D.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structural consequences of turnover-induced homocitrate loss in nitrogenase.
Nat Commun, 14, 2023
|
|
8Q5T
 
 | |
1H1L
 
 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, NIFV MUTANT | | Descriptor: | CHLORIDE ION, CITRIC ACID, FE(8)-S(7) CLUSTER, ... | | Authors: | Mayer, S.M, Gormal, C.A, Smith, B.E, Lawson, D.M. | | Deposit date: | 2002-07-18 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis of the Mofe Protein of Nitrogenase from a Nifv Mutant of Klebsiella Pneumoniae Identifies Citrate as a Ligand to the Molybdenum of Iron Molybdenum Cofactor (Femoco).
J.Biol.Chem., 277, 2002
|
|
8E3U
 
 | |
8E3T
 
 | |
8E3V
 
 | |
1G5P
 
 | | NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Strop, P, Takahara, P.M, Chiu, H.J, Angove, H.C, Burgess, B.K, Rees, D.C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the all-ferrous [4Fe-4S]0 form of the nitrogenase iron protein from Azotobacter vinelandii.
Biochemistry, 40, 2001
|
|
6O7M
 
 | | Nitrogenase MoFeP mutant F99Y from Azotobacter vinelandii in the indigo carmine oxidized state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
6O7L
 
 | |
6O7N
 
 | |
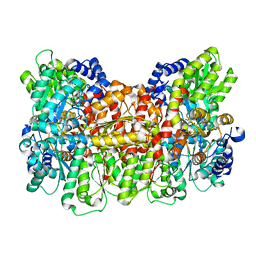
6O7Q
 
 | |
6O7S
 
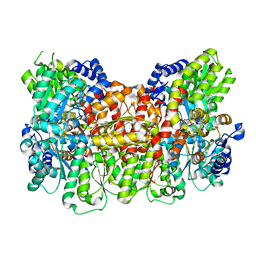 | |
6O7P
 
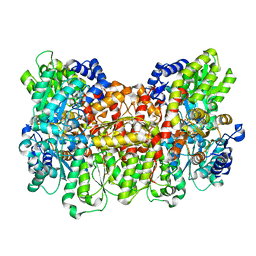 | | Nitrogenase MoFeP mutant F99Y from Azotobacter vinelandii in the dithionite reduced state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
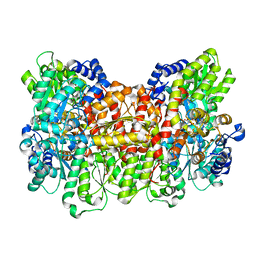
6O7O
 
 | |
6O7R
 
 | | Nitrogenase MoFeP mutant F99Y, S188A from Azotobacter vinelandii in the dithionite reduced state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
6NZJ
 
 | | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO2 Capture by a Surface-Exposed [Fe4S4] Cluster | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein, SULFATE ION | | Authors: | Rettberg, L.A, Kang, W, Stiebritz, M.T, Hiller, C.J, Lee, C.C, Liedtke, J, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of a Nitrogenase Iron Protein from Methanosarcina acetivorans: Implications for CO 2 Capture by a Surface-Exposed [Fe 4 S 4 ] Cluster.
Mbio, 10, 2019
|
|
7AIZ
 
 | | Vanadium nitrogenase VFe protein, high CO state | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Einsle, O. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two ligand-binding sites in CO-reducing V nitrogenase reveal a general mechanistic principle.
Sci Adv, 7, 2021
|
|
