2MEF
 
 | | CONTRIBUTION OF HYDROPHOBIC EFFECT TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1998-05-04 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of amino acid substitutions at two different interior positions to the conformational stability of human lysozyme
Protein Eng., 12, 1999
|
|
6IOQ
 
 | | The ligand binding domain of Mlp24 with glycine | | Descriptor: | CALCIUM ION, GLYCINE, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOU
 
 | | The ligand binding domain of Mlp24 with serine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
4EST
 
 | | CRYSTAL STRUCTURE OF THE COVALENT COMPLEX FORMED BY A PEPTIDYL ALPHA,ALPHA-DIFLUORO-BETA-KETO AMIDE WITH PORCINE PANCREATIC ELASTASE AT 1.78-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, ELASTASE, INHIBITOR ACE-ALA-PRO-VAI-DIFLUORO-N-PHENYLETHYLACETAMIDE, ... | | Authors: | Takahashi, L.H, Radhakrishnan, R, Rosenfieldjunior, R.E, Meyerjunior, E.F, Trainor, D.A. | | Deposit date: | 1989-05-15 | | Release date: | 1992-04-15 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Covalent Complex Formed by a Peptidyl Alpha,Alpha-Difluoro-Beta-Keto Amide with Porcine Pancreatic Elastase at 1.78-Angstroms Resolution
J.Am.Chem.Soc., 111, 1989
|
|
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OI7
 
 | | The Crystal Structure of Succinyl-CoA synthetase alpha subunit from Thermus Thermophilus | | Descriptor: | SUCCINYL-COA SYNTHETASE ALPHA CHAIN | | Authors: | Takahashi, H, Tokunaga, Y, Kuroishi, C, Babayeva, N, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The Crystal Structure of Succinyl-Coa Synthetase from Thermus Thermophilus
To be Published
|
|
5EST
 
 | | Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate | | Descriptor: | CALCIUM ION, ELASTASE, N~2~-[(benzyloxy)carbonyl]-N-[(1R,2S)-1-(dihydroxyboranyl)-2-methylbutyl]-L-alaninamide, ... | | Authors: | Takahashi, L.H, Radhakrishnan, R, Rosenfieldjunior, R.E, Meyerjunior, E.F. | | Deposit date: | 1989-05-15 | | Release date: | 1992-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic analysis of the inhibition of porcine pancreatic elastase by a peptidyl boronic acid: structure of a reaction intermediate.
Biochemistry, 28, 1989
|
|
3AI7
 
 | | Crystal Structure of Bifidobacterium Longum Phosphoketolase | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | | Authors: | Takahashi, K, Tagami, U, Shimba, N, Kashiwagi, T, Ishikawa, K, Suzuki, E. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bifidobacterium Longum phosphoketolase; key enzyme for glucose metabolism in Bifidobacterium
Febs Lett., 584, 2010
|
|
2LAB
 
 | |
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
6IOS
 
 | | The ligand binding domain of Mlp24 with proline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IIE
 
 | | Crystal structure of human diacylglycerol kinase alpha EF-hand domains bound to Ca2+ | | Descriptor: | CALCIUM ION, Diacylglycerol kinase alpha, GLYCEROL, ... | | Authors: | Takahashi, D, Suzuki, K, Sakamoto, T, Iwamoto, T, Murata, T, Sakane, F. | | Deposit date: | 2018-10-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure and calcium-induced conformational changes of diacylglycerol kinase alpha EF-hand domains.
Protein Sci., 28, 2019
|
|
6IOV
 
 | | The ligand binding domain of Mlp37 with arginine | | Descriptor: | ARGININE, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
6IOT
 
 | | The ligand binding domain of Mlp24 with arginine | | Descriptor: | ARGININE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOR
 
 | | The ligand binding domain of Mlp24 with asparagine | | Descriptor: | ASPARAGINE, CALCIUM ION, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
8DXR
 
 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 5 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXP
 
 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 3 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXQ
 
 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 4 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXO
 
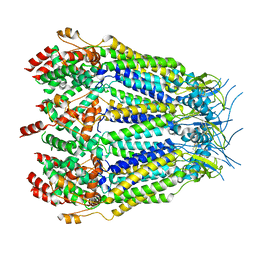 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 2 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
8DXN
 
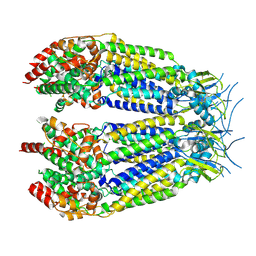 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 1 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
6L88
 
 | |
2YSO
 
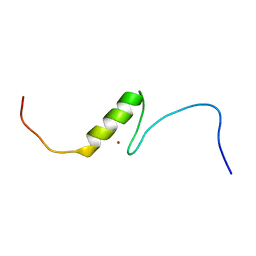 | | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog
To be Published
|
|
2RQA
 
 | | Solution structure of LGP2 CTD | | Descriptor: | ATP-dependent RNA helicase DHX58, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
