8EX3
 
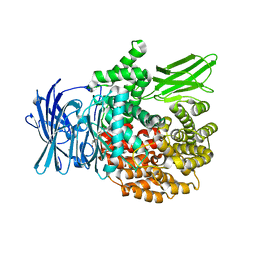 | | Plasmodium falciparum M1 in complex with inhibitor 9aa | | Descriptor: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
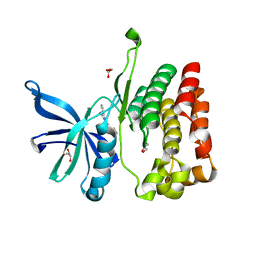
8EX2
 
 | |
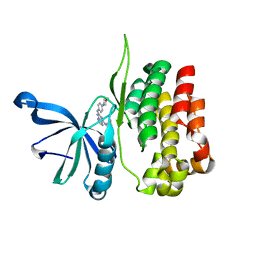
8EX1
 
 | |
8EWZ
 
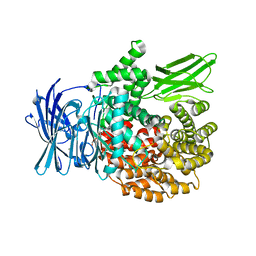 | | Plasmodium falciparum M1 in complex with inhibitor 9c | | Descriptor: | (2R)-2-(cyclopentylcarbamamido)-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EWY
 
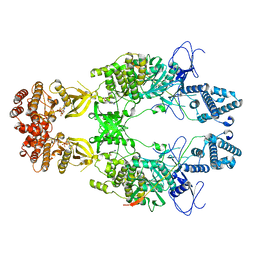 | | Structure of Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | Authors: | Caveney, N.A, Saxton, R.A, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-10-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of Janus kinase trans-activation.
Cell Rep, 42, 2023
|
|
8EWX
 
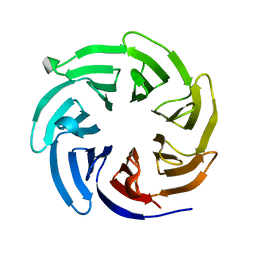 | |
8EWV
 
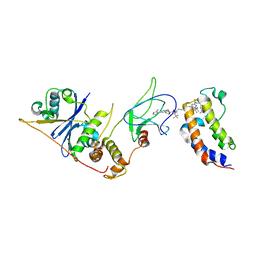 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
8EWT
 
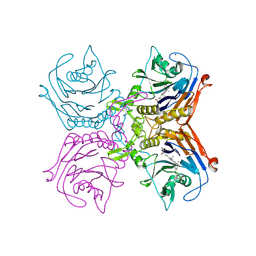 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-{(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-[(prop-2-yn-1-yl)oxy]hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl}-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8EWS
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-(pyridin-4-yl)propanamide}bis[2-(quinolin-2-yl-kappaN)-1-benzothiophen-3-yl-kappaC~3~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWR
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWQ
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]benzamide}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWP
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-(pyridin-4-yl)propanamide}bis[2-(quinolin-2-yl-kappaN)-1-benzothiophen-3-yl-kappaC~3~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWO
 
 | | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, PA1813, ... | | Authors: | Stogios, P.J, Skarina, T, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa
To Be Published
|
|
8EWN
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-(pyridin-4-yl)propanamide}bis[3,5-difluoro-2-(pyridin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWM
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methylidene]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[3,5-difluoro-2-(pyridin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWL
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {N-[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]-3-(pyridin-4-yl)propanamide}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWG
 
 | | Cryo-EM structure of a riboendonclease | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | Authors: | Li, Z, Wang, F. | | Deposit date: | 2022-10-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
8EWF
 
 | |
8EWE
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EWD
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, {tert-butyl [1-{[([2,2'-bipyridin]-5-yl-kappa~2~N~1~,N~1'~)methyl]amino}-1-oxo-3-(pyridin-4-yl)propan-2-yl]carbamate}bis[2-(quinolin-2-yl-kappaN)phenyl-kappaC~1~]iridium(1+) | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2022-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Ir(III) Photosensors for the Major Human Drug-Metabolizing Enzyme Cytochrome P450 3A4.
Inorg.Chem., 62, 2023
|
|
8EW5
 
 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
8EW4
 
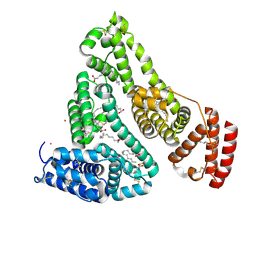 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Unciano, J, Lea, K, Kim, L, Lenkiewicz, J, Starban, I, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EW1
 
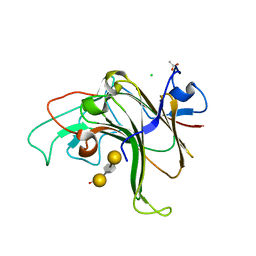 | | Structure of Bacple_01703-E145L | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
8EVR
 
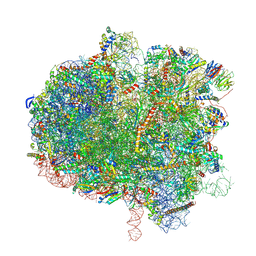 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure II | | Descriptor: | (1S,3S,3aR,4S,4aR,7R,7aR,8aS)-8a-{[(6-deoxy-4-O-methyl-alpha-D-altropyranosyl)oxy]methyl}-4-formyl-7-methyl-3-(propan-2-yl)decahydro-1,4-methano-s-indacene-3a(1H)-carboxylate, 18S rRNA, 25S rRNA, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
