1XT4
 
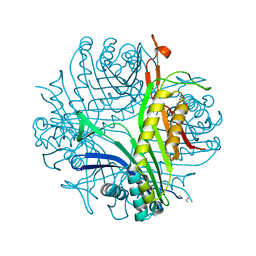 | | Urate Oxidase From Aspergillus Flavus Complexed With Guanine | | Descriptor: | GUANINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-10-21 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1R4U
 
 | |
1R4S
 
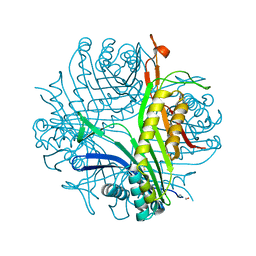 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 9-METHYL URIC ACID | | Descriptor: | 9-METHYL URIC ACID, CYSTEINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R51
 
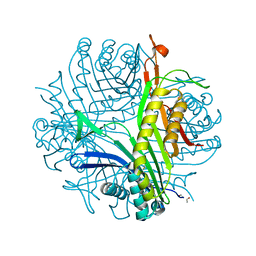 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 8-AZAXANTHIN | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Prange, T, Retailleau, P, Colloc'h, N. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SK9
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A, PHOSPHATE ION | | Authors: | Liu, Q, Huang, Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
1SKA
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A | | Authors: | Liu, Q, Huang, Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
1SK8
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A, PHOSPHATE ION | | Authors: | Liu, Q, Huang, Q.Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2005-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
1SKB
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A | | Authors: | Liu, Q, Huang, Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
6R4E
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate and L-Glu | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4I
 
 | | Crystal structure of human GFAT-1 G461E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4H
 
 | | Crystal structure of human GFAT-1 G451E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4J
 
 | | Crystal structure of human GFAT-1 G451E in complex with UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4F
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4G
 
 | | Crystal structure of human GFAT-1 in complex with UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, MAGNESIUM ION, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6SVP
 
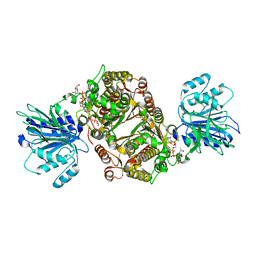 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
5D21
 
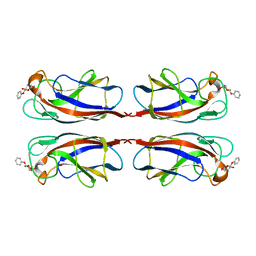 | | Multivalency Effects in Glycopeptide Dendrimer Inhibitors of Pseudomonas aeruginosa Biofilms Targeting Lectin LecA | | Descriptor: | CALCIUM ION, LecA, phenyl beta-D-galactopyranoside | | Authors: | Bergmann, M, Michaud, G, Visini, R, Jin, X, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalency effects on Pseudomonas aeruginosa biofilm inhibition and dispersal by glycopeptide dendrimers targeting lectin LecA.
Org.Biomol.Chem., 14, 2016
|
|
3NGA
 
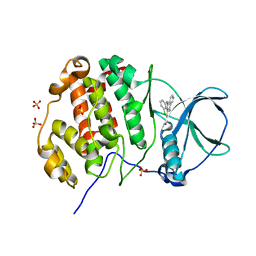 | | Human CK2 catalytic domain in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2010-06-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis of CX-4945 binding to human protein kinase CK2.
Febs Lett., 585, 2011
|
|
4JK8
 
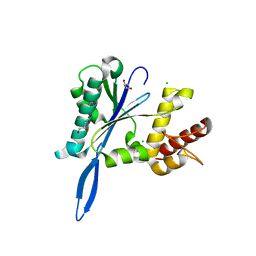 | | Open and closed forms of R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKF
 
 | | Open and closed forms of T1791P+R1865A human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKA
 
 | | Open and closed forms of R1865A human PRP8 RNase H-like domain with bound Co ion | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKE
 
 | | Open and closed forms of T1789P human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LE2
 
 | |
4LE1
 
 | | Crystal structure of the receiver domain of DesR in the inactive state | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4LE0
 
 | | Crystal structure of the receiver domain of DesR in complex with beryllofluoride and magnesium | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4LDZ
 
 | | Crystal structure of the full-length response regulator DesR in the active state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
