1EAN
 
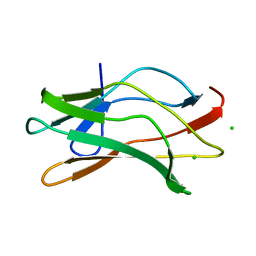 | | THE RUNX1 Runt domain at 1.70A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-13 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1GQG
 
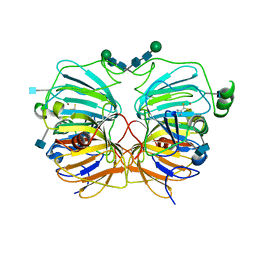 | | Quercetin 2,3-dioxygenase in complex with the inhibitor diethyldithiocarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase.1.Ligand-Induced Coordination Changes Probed by X-Ray Crystallography: Inhibition, Ordering Effect and Mechanistic Insights
Biochemistry, 41, 2002
|
|
1GYU
 
 | |
1GWA
 
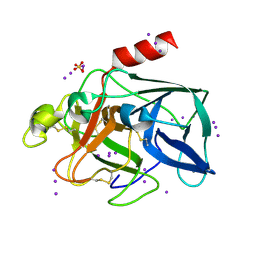 | | Triiodide derivative of porcine pancreas elastase | | Descriptor: | CALCIUM ION, ELASTASE 1, IODIDE ION, ... | | Authors: | Evans, G, Bricogne, G. | | Deposit date: | 2002-03-13 | | Release date: | 2002-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triiodide Derivatization and Combinatorial Counter-Ion Replacement: Two Methods for Enhancing Phasing Signal Using Laboratory Cu Kalpha X-Ray Equipment
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1AV2
 
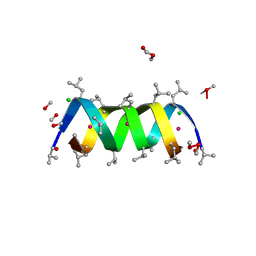 | | Gramicidin A/CsCl complex, active as a dimer | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Burkhart, B.M, Li, N, Langs, D.A, Duax, W.L. | | Deposit date: | 1997-09-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Conducting Form of Gramicidin a is a Right-Handed Double-Stranded Double Helix.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1GXA
 
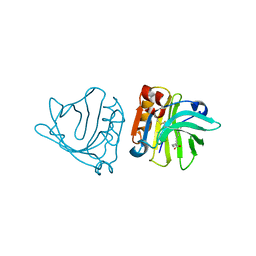 | |
1GRW
 
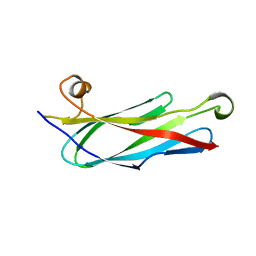 | | C. elegans major sperm protein | | Descriptor: | MAJOR SPERM PROTEIN 31/40/142 | | Authors: | Baker, A.M.E, Roberts, T.M, Stewart, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A Resolution Crystal Structure of Helices of the Motile Major Sperm Protein (Msp) of Caenorhabditis Elegans
J.Mol.Biol., 319, 2002
|
|
1FT0
 
 | | CRYSTAL STRUCTURE OF TRUNCATED HUMAN RHOGDI K113A MUTANT | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Longenecker, K.L, Garrard, S.M, Sheffield, P.J, Derewenda, Z.S. | | Deposit date: | 2000-09-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1F8T
 
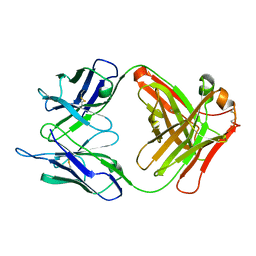 | | FAB (LNKB-2) OF MONOCLONAL ANTIBODY, CRYSTAL STRUCTURE | | Descriptor: | ANTIBODY FAB FRAGMENT (HEAVY CHAIN), ANTIBODY FAB FRAGMENT (LIGHT CHAIN) | | Authors: | Fokin, A.V, Afonin, P.V, Mikhailova, I.I, Tsygannik, I.N, Mareeva, T.I. | | Deposit date: | 2000-07-05 | | Release date: | 2001-07-05 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spatial structure of a Fab-fragment of a monoclonal antibody to human interleukin-2 in two crystalline forms at a resolution of 2.2 and 2.9 angstroms
Bioorg.Khim., 26, 2000
|
|
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1D27
 
 | |
1D65
 
 | |
1D82
 
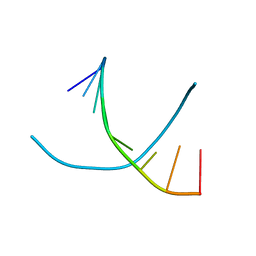 | |
1D21
 
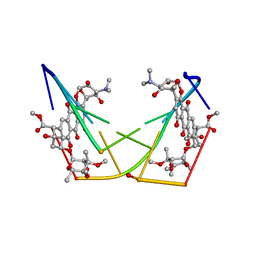 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
1D17
 
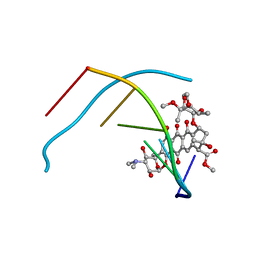 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
1FQ1
 
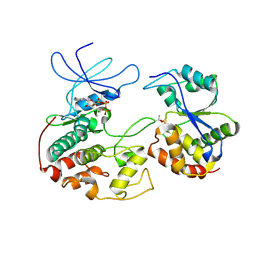 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1D58
 
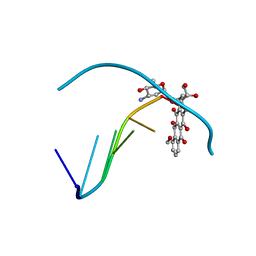 | | THE MOLECULAR STRUCTURE OF A 4'-EPIADRIAMYCIN COMPLEX WITH D(TGATCA) AT 1.7 ANGSTROM RESOLUTION-COMPARISON WITH THE STRUCTURE OF 4'-EPIADRIAMYCIN D(TGTACA) AND D(CGATCG) COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*TP*GP*AP*TP*CP*A)-3') | | Authors: | Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-02-20 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of a 4'-epiadriamycin complex with d(TGATCA) at 1.7A resolution: comparison with the structure of 4'-epiadriamycin d(TGTACA) and d(CGATCG) complexes.
Nucleic Acids Res., 20, 1992
|
|
1D92
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH G.T MISMATCHED BASE-PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*TP*CP*C)-3') | | Authors: | Hunter, W.N, Kneale, G, Brown, T, Rabinovich, D, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs.
J.Mol.Biol., 190, 1986
|
|
1FK0
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FKN
 
 | | Structure of Beta-Secretase Complexed with Inhibitor | | Descriptor: | MEMAPSIN 2, inhibitor | | Authors: | Hong, L, Koelsch, G, Lin, X, Wu, S, Terzyan, S, Ghosh, A, Zhang, X.C, Tang, J. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor.
Science, 290, 2000
|
|
1FOC
 
 | | Cytochrome C557: improperly folded thermus thermophilus C552 | | Descriptor: | CYTOCHROME RC557, HEME C | | Authors: | McRee, D.E, Williams, P.A, Fee, J.A, Bren, K.L. | | Deposit date: | 2000-08-27 | | Release date: | 2000-11-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recombinant cytochrome rC557 obtained from Escherichia coli cells expressing a truncated Thermus thermophilus cycA gene. Heme inversion in an improperly matured protein
J.Biol.Chem., 276, 2001
|
|
1GXM
 
 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | GLYCEROL, PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GXO
 
 | | Mutant D189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid | | Descriptor: | CALCIUM ION, PECTATE LYASE, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GXV
 
 | |
1D16
 
 | |
