1Z7T
 
 | |
1UHZ
 
 | | Solution structure of dsRNA binding domain in Staufen homolog 2 | | Descriptor: | staufen (RNA binding protein) homolog 2 | | Authors: | He, F, Muto, Y, Obayashi, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Koboyashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dsRNA binding domain in Staufen homolog 2
To be Published
|
|
1LKN
 
 | |
1YZM
 
 | | Structure of Rabenosyn (458-503), Rab4 binding domain | | Descriptor: | FYVE-finger-containing Rab5 effector protein rabenosyn-5 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1UIY
 
 | | Crystal Structure of Enoyl-CoA Hydratase from Thermus Thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Enoyl-CoA Hydratase, GLYCEROL | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-24 | | Release date: | 2003-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
1Z0D
 
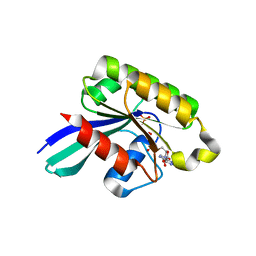 | | GDP-Bound Rab5c GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1UWR
 
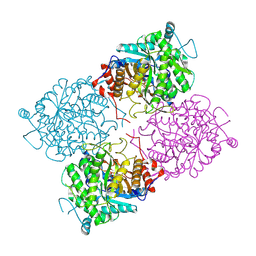 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Studies of the Beta-Glycosidase from Sulfolobus Solfataricus in Complex with Covalently and Noncovalently Bound Inhibitors.
Biochemistry, 43, 2004
|
|
1UJM
 
 | |
1UIL
 
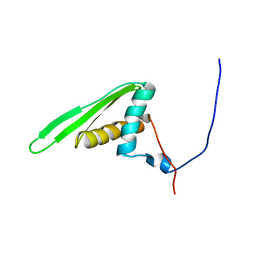 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | Descriptor: | Double-stranded RNA-binding motif | | Authors: | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
1ZOQ
 
 | | IRF3-CBP complex | | Descriptor: | CREB-binding protein, Interferon regulatory factor 3 | | Authors: | Qin, B, Lin, K. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of IRF-3 in complex with CBP.
Structure, 13, 2005
|
|
1ZP8
 
 | | HIV Protease with inhibitor AB-2 | | Descriptor: | Pol polyprotein, [1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL (1R,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLCARBAMATE | | Authors: | Brik, A, Alexandratos, J.N, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors.
Chembiochem, 6, 2005
|
|
1UJS
 
 | |
1UJR
 
 | | Solution structure of WWE domain in BAB28015 | | Descriptor: | hypothetical protein AK012080 | | Authors: | He, F, Muto, Y, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-11 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in BAB28015
To be Published
|
|
1UJY
 
 | | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6 | | Descriptor: | Rho guanine nucleotide exchange factor 6 | | Authors: | He, F, Muto, Y, Uda, H, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-02-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3 domain in Rac/Cdc42 guanine nucleotide exchange factor(GEF) 6
To be Published
|
|
1LRM
 
 | |
1UJU
 
 | |
1UJT
 
 | |
1UJX
 
 | |
2A0Y
 
 | | Structure of human purine nucleoside phosphorylase H257D mutant | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Murkin, A.S, Shi, W, Schramm, V.L. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
1UYL
 
 | | Structure-Activity Relationships in purine-based inhibitor binding to HSP90 isoforms | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
2A47
 
 | | Crystal structure of amFP486 H199T | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ULT
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, long chain fatty acid-CoA ligase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of the substrate specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer
J.Biol.Chem., 279, 2004
|
|
1V4H
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima F52A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1UMV
 
 | | Crystal structure of an acidic, non-myotoxic phospholipase A2 from the venom of Bothrops jararacussu | | Descriptor: | CALCIUM ION, HYPOTENSIVE PHOSPHOLIPASE A2 | | Authors: | Murakami, M.T, Watanabe, L, Cintra, A.C.O, Arni, R.K. | | Deposit date: | 2003-08-28 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of an Acidic Platelet Aggregation Inhibitor and Hypotensive Phospholipase A(2) in the Monomeric and Dimeric States: Insights Into its Oligomeric State
Biochem.Biophys.Res.Commun., 323, 2004
|
|
2A89
 
 | | Monomeric Sarcosine Oxidase: Structure of a covalently flavinylated amine oxidizing enzyme | | Descriptor: | (N5,C4A)-(ALPHA-HYDROXY-PROPANO)-3,4,4A,5-TETRAHYDRO-FLAVIN-ADENINE DINUCLEOTIDE, CHLORIDE ION, Monomeric sarcosine oxidase, ... | | Authors: | Chen, Z.-W, Zhao, G, Martinovic, S, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2005-07-07 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the sodium borohydride-reduced N-(cyclopropyl)glycine adduct of the flavoenzyme monomeric sarcosine oxidase.
Biochemistry, 44, 2005
|
|
