2XEC
 
 | | Nocardia farcinica maleate cis-trans isomerase bound to TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PUTATIVE MALEATE ISOMERASE | | Authors: | Fisch, F, Martinez-Fleites, C, Baudendistel, N, Hauer, B, Turkenburg, J.P, Hart, S, Bruce, N.C, Grogan, G. | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Covalent Succinylcysteine-Like Intermediate in the Enzyme-Catalyzed Transformation of Maleate to Fumarate by Maleate Isomerase.
J.Am.Chem.Soc., 132, 2010
|
|
2XOD
 
 | | Crystal structure of flavoprotein NrdI from Bacillus anthracis in the oxidised form | | Descriptor: | CACODYLATE ION, FLAVIN MONONUCLEOTIDE, NRDI PROTEIN, ... | | Authors: | Johansson, R, Sprenger, J, Torrents, E, Sahlin, M, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2010-08-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | High Resolution Crystal Structures of Nrdi in the Oxidised and Reduced States: An Unusual Flavodoxin
FEBS J., 277, 2010
|
|
2HQU
 
 | | Human dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Varga, B, Vertessy, B.G. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase.
Febs Lett., 581, 2007
|
|
2XR0
 
 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
2XSX
 
 | | Crystal structure of human beta enolase ENOB | | Descriptor: | 1,2-ETHANEDIOL, BETA-ENOLASE, MAGNESIUM ION, ... | | Authors: | Vollmar, M, Krysztofinska, E, Chaikuad, A, Krojer, T, Cocking, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Beta Enolase Enob
To be Published
|
|
2XOE
 
 | | Crystal structure of flavoprotein NrdI from Bacillus anthracis in the semiquinone form | | Descriptor: | ACETATE ION, CACODYLATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, R, Sprenger, J, Torrents, E, Sahlin, M, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2010-08-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Crystal Structures of Nrdi in the Oxidised and Reduced States: An Unusual Flavodoxin
FEBS J., 277, 2010
|
|
2XQQ
 
 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide (Ac-SRGTQTE). | | Descriptor: | ACETATE ION, DYNEIN LIGHT CHAIN 2, CYTOPLASMIC, ... | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-09-07 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Directed Evolution Reveals the Binding Motif Preference of the Lc8/Dynll Hub Protein and Predicts Large Numbers of Novel Binders in the Human Proteome
Plos One, 6, 2011
|
|
2WBP
 
 | | Crystal structure of VioC in complex with Fe(II), (2S,3S)- hydroxyarginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ARGININE, FE (II) ION, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2VPA
 
 | | High resolution crystal structure of the antibiotic resistance protein NimA from Deinococcus radiodurans | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Tedesco, C, McSweeney, S.M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-Resolution Structure of the Antibiotic Resistance Protein Nima from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VQ1
 
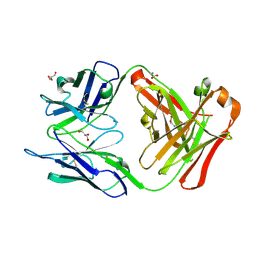 | | anti trimeric Lewis X Fab54-5C10-A | | Descriptor: | ANTI-HUMAN FC GAMMA RECEPTOR III 3G8 GAMMA HEAVY CHAIN VARIABLE REGION, AZIDE ION, GLYCEROL, ... | | Authors: | de Geus, D.C, van Roon, A.M.M, Thomassen, E.A.J, Hokke, C.H, Deelder, A.M, Abrahams, J.P. | | Deposit date: | 2008-03-10 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a Diagnostic Fab Fragment Binding Trimeric Lewis X.
Proteins, 76, 2009
|
|
2FP8
 
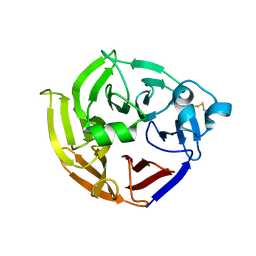 | |
2G1A
 
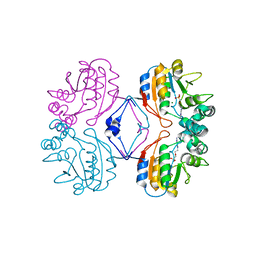 | | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase | | Descriptor: | Class B acid phosphatase, MAGNESIUM ION, {[2-(6-AMINO-9H-PURIN-9-YL)ETHOXY]METHYL}PHOSPHONIC ACID | | Authors: | Leone, R, Calderone, V, Cappelletti, E, Benvenuti, M, Mangani, S. | | Deposit date: | 2006-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between Apha class B acid phosphatase/phosphotransferase
To be published
|
|
2WD7
 
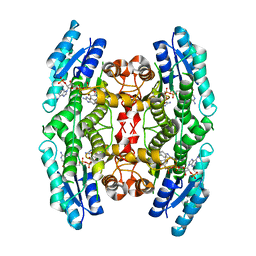 | | PTERIDINE REDUCTASE 1 (PTR1) FROM TRYPANOSOMA BRUCEI IN COMPLEX WITH NADP AND DDD00066750 | | Descriptor: | 6-chloro-1H-benzimidazol-2-amine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Robinson, D.A, Wyatt, P.G, Spinks, D, Brenk, R. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | One Scaffold, Three Binding Modes: Novel and Selective Pteridine Reductase 1 Inhibitors Derived from Fragment Hits Discovered by Virtual Screening.
J.Med.Chem., 52, 2009
|
|
2FR2
 
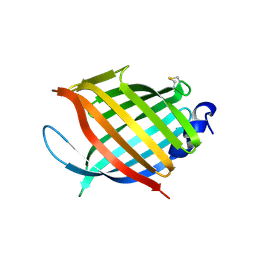 | |
2FZN
 
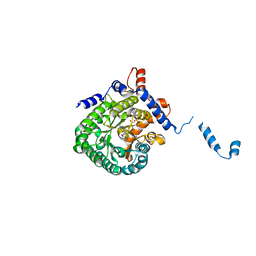 | |
2FPC
 
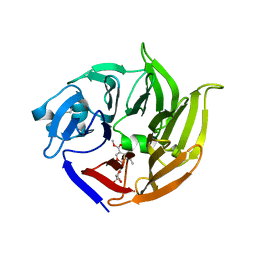 | |
2VYN
 
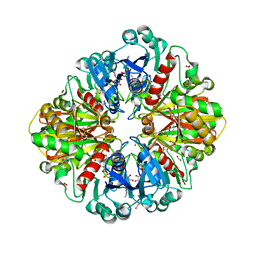 | | Structure of E.Coli GAPDH Rat Sperm GAPDH heterotetramer | | Descriptor: | FORMIC ACID, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Frayne, J, Taylor, A, Hall, L, Hadfield, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Insoluble Rat Sperm Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) Via Heterotetramer Formation with Escherichia Coli Gapdh Reveals Target for Contraceptive Design.
J.Biol.Chem., 284, 2009
|
|
2VYV
 
 | | Structure of E.Coli GAPDH Rat Sperm GAPDH heterotetramer | | Descriptor: | FORMIC ACID, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Frayne, J, Taylor, A, Hall, L, Hadfield, A. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of Insoluble Rat Sperm Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) Via Heterotetramer Formation with Escherichia Coli Gapdh Reveals Target for Contraceptive Design.
J.Biol.Chem., 284, 2009
|
|
2ZUS
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase | | Descriptor: | Lacto-N-biose phosphorylase, MAGNESIUM ION | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZK7
 
 | | Structure of a C-terminal deletion mutant of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) | | Descriptor: | Glucose 1-dehydrogenase related protein | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2008-03-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | C-terminal tail derived from the neighboring subunit is critical for the activity of Thermoplasma acidophilum D-aldohexose dehydrogenase
Proteins, 74, 2009
|
|
2Z5U
 
 | |
2HTO
 
 | | Ruthenium hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), RUTHENIUM (III) HEXAAMINE ION | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2ZUU
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZTV
 
 | | The binary complex of D-3-hydroxybutyrate dehydrogenase with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZUV
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc, Ethylene glycol, and nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
