6YZY
 
 | | Crystal structure of the M295V variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase, SULFATE ION | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
9HIS
 
 | |
6ZHG
 
 | | Ca2+-ATPase from Listeria Monocytogenes in complex with AlF | | Descriptor: | Calcium-transporting ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
7NLN
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with N-acetyl-glutamate | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6O7V
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6OA8
 
 | | Superfolder Green Fluorescent Protein with 4-cyano-L-phenylalanine at the chromophore (position 66) | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION, ... | | Authors: | Piacentini, J, Olenginski, G.M, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
8UTK
 
 | | IL-23R minibinder - 23R-B04dslf02IB | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 23R-B04dslf02IB, ... | | Authors: | Bera, A.K, Berger, S.A, Kang, A, Baker, D. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Preclinical proof of principle for orally delivered Th17 antagonist miniproteins.
Cell, 187, 2024
|
|
9HJ3
 
 | |
6ZBZ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3,4-dimethylphenyl)sulfonylamino]-5-fluoranyl-benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC3
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-2-(methylsulfonylamino)benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
8UQ7
 
 | | Alzheimer's disease PHF complexed with PET ligand MK-6240 | | Descriptor: | 6-fluoro-3-(1H-pyrrolo[2,3-c]pyridin-1-yl)isoquinolin-5-amine, Microtubule-associated protein tau | | Authors: | Kunach, P, Shahmoradian, S.H, Diamond, M.I, Rosa-Neto, P. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Cryo-EM structure of Alzheimer's disease tau filaments with PET ligand MK-6240.
Nat Commun, 15, 2024
|
|
8J1C
 
 | |
8J1G
 
 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
6RTW
 
 | | Crystal structure of the Patched-1 (PTCH1) ectodomain in complex with nanobody NB64 and cholesterol-hemisuccinate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Llama-derived nanobody NB64, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6Z1K
 
 | |
6Z1L
 
 | |
6QCN
 
 | | Human Sirt2 in complex with ADP-ribose and the inhibitor quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Riemer, S, You, W, Steegborn, C. | | Deposit date: | 2018-12-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives.
Sci Rep, 9, 2019
|
|
6F1S
 
 | | C-terminal domain of CglI restriction endonuclease H subunit | | Descriptor: | 1,2-ETHANEDIOL, CglIIR protein, FORMIC ACID | | Authors: | Tamulaitiene, G, Grigaitis, R, Zaremba, M, Silanskas, A. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The H-subunit of the restriction endonuclease CglI contains a prototype DEAD-Z1 helicase-like motor.
Nucleic Acids Res., 46, 2018
|
|
6ZK3
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
8IXV
 
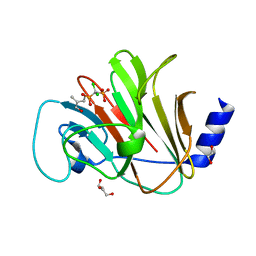 | | Crystal structure of intracellular B30.2 domain of BTN3A in complex with 2Cl-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6ZK4
 
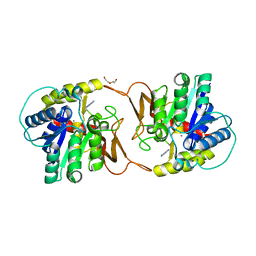 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
5JNW
 
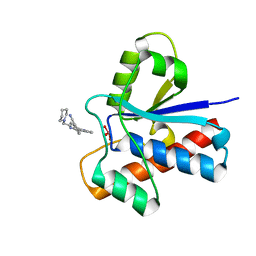 | | Crystal structure of bovine low molecular weight protein tyrosine phosphatase (LMPTP) mutant (W49Y N50E) complexed with vanadate and uncompetitive inhibitor | | Descriptor: | 2-(4-{[3-(piperidin-1-yl)propyl]amino}quinolin-2-yl)benzonitrile, Low molecular weight phosphotyrosine protein phosphatase, VANADATE ION | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
6ZMN
 
 | |
6QE4
 
 | | Re-refinement of 5OLI human IBA57-I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L, Nasta, V. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house high-energy-remote SAD phasing using the magic triangle: how to tackle the P1 low symmetry using multiple orientations of the same crystal of human IBA57 to increase the multiplicity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5IUI
 
 | | Crystal Structure of Anaplastic Lyphoma Kinase (ALK) in complex with 4 | | Descriptor: | ALK tyrosine kinase receptor, N-[3-(4-amino-3-methylphenyl)-1H-pyrazol-5-yl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide | | Authors: | Tu, C.H, Wu, S.Y. | | Deposit date: | 2016-03-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Pyrazolylamine Derivatives Reveal the Conformational Switching between Type I and Type II Binding Modes of Anaplastic Lymphoma Kinase (ALK).
J.Med.Chem., 59, 2016
|
|
