8F97
 
 | | Compound 5 bound to procaspase-6 | | Descriptor: | 1,2-ETHANEDIOL, 2-({[(2M)-[2,3'-bipyridin]-2'-yl]amino}methyl)-5-fluorophenol, CHLORIDE ION, ... | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8DDF
 
 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
6NE6
 
 | |
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
8Z6X
 
 | |
8WBE
 
 | |
8F39
 
 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6O24
 
 | | Crystal structure of 4498 Fab in complex with circumsporozoite protein NANP3 and anti-Kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4498 Fab heavy chain, 4498 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
8K12
 
 | | SID1 transmembrane family member 2 | | Descriptor: | SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
6WN6
 
 | | Crystal structure of 3-keto-D-glucoside 4-epimerase, YcjR, from E. coli, apo form | | Descriptor: | 1,2-ETHANEDIOL, 3-keto-D-glucoside 4-epimerase, MANGANESE (II) ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Mukherjee, K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Reaction Mechanism of YcjR, an Epimerase That Facilitates the Interconversion of d-Gulosides to d-Glucosides inEscherichia coli.
Biochemistry, 59, 2020
|
|
6NOF
 
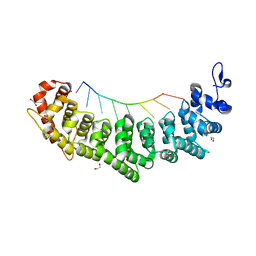 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
1F9O
 
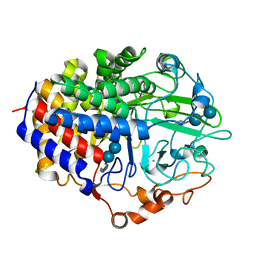 | | Crystal structure of the cellulase Cel48F from C. Cellulolyticum with the thiooligosaccharide inhibitor PIPS-IG3 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-iodophenyl 1,4-dithio-beta-D-glucopyranoside | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
8DOG
 
 | | Dehaloperoxidase B in complex with Bisphenol E | | Descriptor: | 4-[1-(4-hydroxyphenyl)ethyl]phenol, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | de Serrano, V.S, Yun, D, Ghiladi, R. | | Deposit date: | 2022-07-13 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Oxidation of bisphenol A (BPA) and related compounds by the multifunctional catalytic globin dehaloperoxidase.
J.Inorg.Biochem., 238, 2023
|
|
1FGG
 
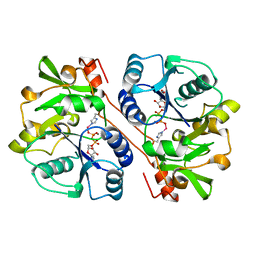 | | CRYSTAL STRUCTURE OF 1,3-GLUCURONYLTRANSFERASE I (GLCAT-I) COMPLEXED WITH GAL-GAL-XYL, UDP, AND MN2+ | | Descriptor: | GLUCURONYLTRANSFERASE I, MANGANESE (II) ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pedersen, L.C, Tsuchida, K, Kitagawa, H, Sugahara, K, Darden, T.A. | | Deposit date: | 2000-07-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I.
J.Biol.Chem., 275, 2000
|
|
8D95
 
 | |
6X35
 
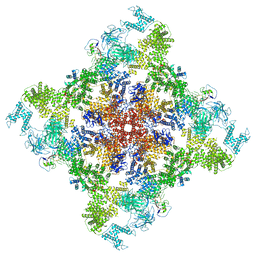 | | Pig R615C RyR1 in complex with CaM, EGTA (class 1, open) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7RBR
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7OQN
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 30 minutes soaking | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
8DNK
 
 | | Crystal structure of human KRAS G12C covalently bound with Taiho WO2020/085493A1 compound 6 | | Descriptor: | 2-{[(5-tert-butyl-6-chloro-1H-indazol-3-yl)amino]methyl}-4-chloro-1-methyl-N-(1-propanoylazetidin-3-yl)-1H-imidazole-5-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7OQM
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.05 Angstroms resolution, 20 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQF
 
 | | Human OMPD-domain of UMPS in complex with OMP at 1.05 Angstrom resolution, 5 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQI
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.15 Angstrom resolution, 10 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OU6
 
 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | Protein O-GlcNAcase, ~{N}-[(3~{Z},6~{S},7~{R},8~{R},8~{a}~{S})-7,8-bis(oxidanyl)-3-(phenylmethyl)imino-1,5,6,7,8,8~{a}-hexahydro-[1,3]thiazolo[3,4-a]pyridin-6-yl]ethanamide | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7OUZ
 
 | |
7OQK
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 15 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
