7M2F
 
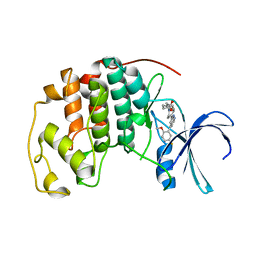 | | CDK2 with compound 14 inhibitor with carboxylate | | Descriptor: | Cyclin-dependent kinase 2, [(1r,4r)-4-{4-[4-(5-fluoro-2-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-3,6-dihydropyridin-1(2H)-yl}cyclohexyl]acetic acid | | Authors: | Longenecker, K.L, Qiu, W, Korepanova, A, Tong, Y. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Balancing Properties with Carboxylates: A Lead Optimization Campaign for Selective and Orally Active CDK9 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
8SPX
 
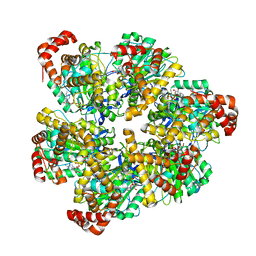 | | PS3 F1 Rotorless, high ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
6SX3
 
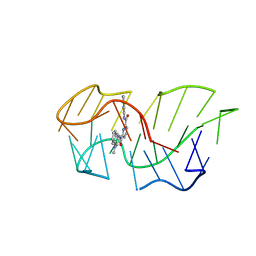 | | Intercalation of heterocyclic ligand between quartets in G-rich tetrahelical structure | | Descriptor: | VK2, ~{N}2,~{N}6-bis(1-methylquinolin-1-ium-3-yl)pyridine-2,6-dicarboxamide | | Authors: | Kotar, A, Kocman, V, Plavec, J. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Intercalation of a Heterocyclic Ligand between Quartets in a G-Rich Tetrahelical Structure.
Chemistry, 26, 2020
|
|
6N5A
 
 | |
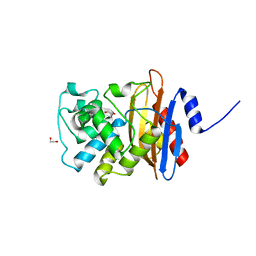
5HX9
 
 | |
6N67
 
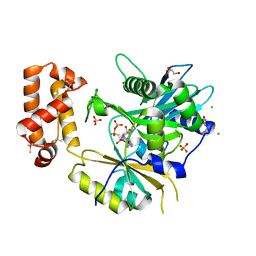 | |
8VPN
 
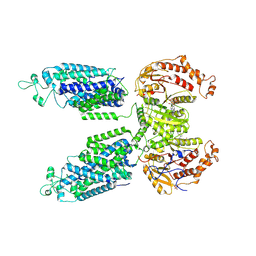 | | Phosphorylated human NCC in complex with indapamide | | Descriptor: | 4-chloro-N-[(2S)-2-methyl-2,3-dihydro-1H-indol-1-yl]-3-sulfamoylbenzamide, ADENOSINE-5'-TRIPHOSPHATE, Solute carrier family 12 member 3 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2024-01-16 | | Release date: | 2024-09-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural bases for Na + -Cl - cotransporter inhibition by thiazide diuretic drugs and activation by kinases.
Nat Commun, 15, 2024
|
|
8EYJ
 
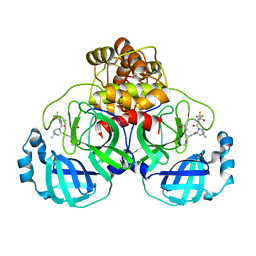 | | Crystal Structure of uncleaved SARS-CoV-2 Main Protease C145S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Nat Commun, 14, 2023
|
|
6W46
 
 | | Valine-Containing Collagen Peptide | | Descriptor: | 1,2-ETHANEDIOL, Collagen-like peptide | | Authors: | Chenoweth, D.M, Melton, S.D. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rules for the design of aza-glycine stabilized triple-helical collagen peptides.
Chem Sci, 11, 2020
|
|
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
7O3M
 
 | | Crystal Structure of AcrB Single Mutant - 1 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 1
To Be Published
|
|
8C80
 
 | | Cryo-EM structure of the yeast SPT-Orm1-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
7QVT
 
 | | BAZ2A bromodomain in complex with picolinamide derivative fragment 13 | | Descriptor: | 1,2-ETHANEDIOL, 4-piperidin-4-yloxypyridine-2-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-23 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
8C81
 
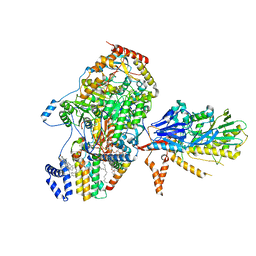 | | Cryo-EM structure of the yeast SPT-Orm1-Sac1 complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C82
 
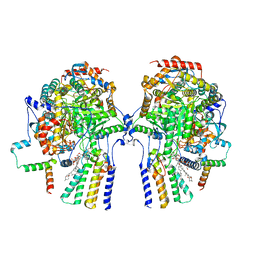 | | Cryo-EM structure of the yeast SPT-Orm1-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8ERX
 
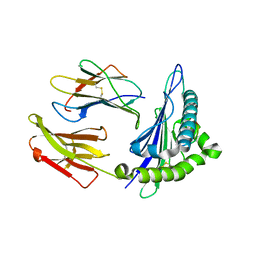 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
6N45
 
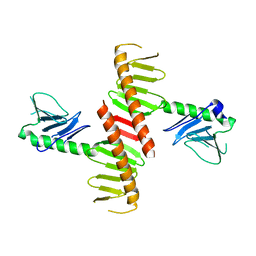 | |
7O83
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(7S)-11-chloro-12-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,15,17-tetrazatetracyclo[8.7.1.02,7.014,18]octadeca-1(17),10,12,14(18),15-pentaen-5-yl]prop-2-en-1-one, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7QT1
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitution e170N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
9FKB
 
 | | Tail of emppty Haloferax tailed virus 1 | | Descriptor: | Baseplate to tube adapter protein gp41, HK97 gp6-like/SPP1 gp15-like head-tail connector, MAGNESIUM ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2024-06-03 | | Release date: | 2025-06-18 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM resolves the structure of the archaeal dsDNA virus HFTV1 from head to tail.
Sci Adv, 11, 2025
|
|
6MO2
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{5-[(piperidin-4-yl)methoxy]-3-[4-(1H-pyrazol-4-yl)phenyl]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
7B6P
 
 | | Crystal structure of E.coli MurE - C269S C340S C450S in complex with Ellman's reagent | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7QSY
 
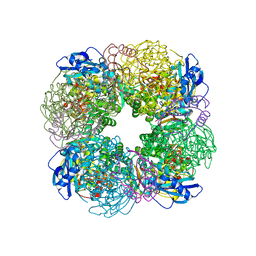 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSZ
 
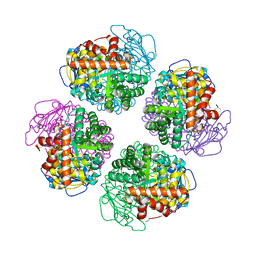 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8 complex with substitution e170N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7OPA
 
 | | Purine nucleoside phosphorylase(DeoD-type) from H. pylori with 6-benzylthiopurine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-benzylthio-2-chloropurine, GLYCEROL, ... | | Authors: | Narczyk, M, Stefanic, Z. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium.
J Enzyme Inhib Med Chem, 37, 2022
|
|
