9PSK
 
 | | In situ MicroED structure of IL-33 activated human eosinophil major basic protein-1 | | Descriptor: | Bone marrow proteoglycan, CHLORIDE ION | | Authors: | Yang, J.E, Bingman, C.A, Mitchell, J, Mosher, D, Wright, E.R. | | Deposit date: | 2025-07-25 | | Release date: | 2025-09-03 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.18 Å) | | Cite: | In situ MicroED structure of the human eosinophil major basic protein-1
To Be Published
|
|
6UXM
 
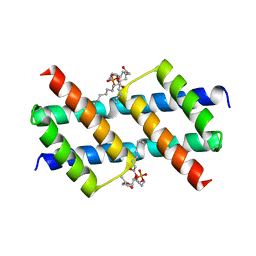 | |
9QBY
 
 | | Yeast 20S proteasome mutant: beta5_G128V (b5-propeptide in trans) in complex with ONX0914 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2025-03-04 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Proteasome-associated autoinflammatory syndromes: The impact of mutations in proteasome subunits on particle assembly, structure, and activity.
Structure, 2025
|
|
8CHW
 
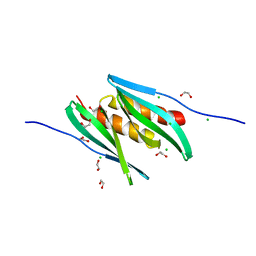 | |
9PDA
 
 | |
8CLU
 
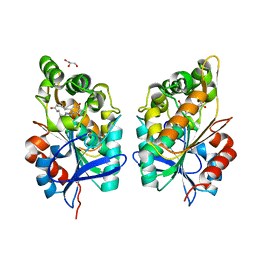 | | Zearalenone lactonase from Rhodococcus erythropolis in complex with zearalactamenone | | Descriptor: | (4~{S})-4-methyl-16,18-bis(oxidanyl)-3-azabicyclo[12.4.0]octadeca-1(18),12,14,16-tetraene-2,8-dione, GLYCEROL, Zearalenone lactonase | | Authors: | Puehringer, D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
9S2V
 
 | | NSP14 IN COMPLEX WITH LIGAND TDI-014925-CL-2 (compound 58) | | Descriptor: | CHLORIDE ION, Guanine-N7 methyltransferase nsp14, IMIDAZOLE, ... | | Authors: | Steinbacher, S, Huggins, D.J. | | Deposit date: | 2025-07-22 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery, Optimization, and Evaluation of Non-Nucleoside SARS-CoV-2 NSP14 Inhibitors.
J.Med.Chem., 68, 2025
|
|
8YK7
 
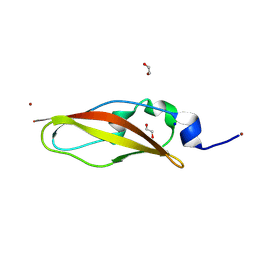 | | Structure of Rib domain from surface adhesin of Limosilactobacillus reuteri | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the Rib domain of the cell-wall-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
1ELK
 
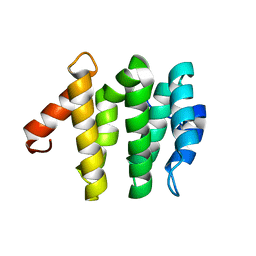 | | VHS domain of TOM1 protein from H. sapiens | | Descriptor: | TARGET OF MYB1 | | Authors: | Misra, S, Beach, B, Hurley, J.H. | | Deposit date: | 2000-03-13 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the VHS domain of human Tom1 (target of myb 1): insights into interactions with proteins and membranes
Biochemistry, 39, 2000
|
|
1E8B
 
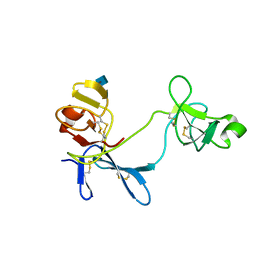 | | Solution structure of 6F11F22F2, a compact three-module fragment of the gelatin-binding domain of human fibronectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FIBRONECTIN | | Authors: | Pickford, A.R, Smith, S.P, Staunton, D, Boyd, J, Campbell, I.D. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The Hairpin Structure of the (6)F1(1)F2(2)F2 Fragment from Human Fibronectin Enhances Gelatin Binding
Embo J., 20, 2001
|
|
7PTI
 
 | |
8Y2R
 
 | | NMR Solution Structure of the 2:1 Coptisine-ATF4-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*TP*AP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*AP*GP*GP*GP*AP*GP*C*(LIG)*(LIG))-3') | | Authors: | Xiao, C.M, Li, Y.P, Liu, Y.S, Dong, R.F, He, X.Y, Lin, Q, Zang, X, Wang, K.B, Xia, Y.Z, Kong, L.Y. | | Deposit date: | 2024-01-27 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | SOLUTION NMR | | Cite: | Overcoming Cancer Persister Cells by Stabilizing the ATF4 Promoter G-quadruplex.
Adv Sci, 11, 2024
|
|
6VGZ
 
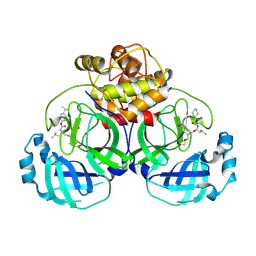 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | Descriptor: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
5A0D
 
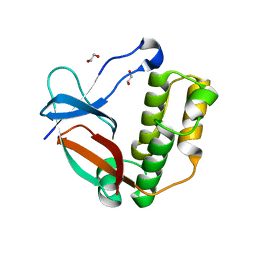 | | N-terminal thioester domain of surface protein from Clostridium perfringens, Cys138Ala mutant | | Descriptor: | 1,2-ETHANEDIOL, SURFACE ANCHORED PROTEIN | | Authors: | Walden, M, Edwards, J.M, Dziewulska, A.M, Kan, S.-Y, Schwarz-Linek, U, Banfield, M.J. | | Deposit date: | 2015-04-17 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An internal thioester in a pathogen surface protein mediates covalent host binding.
Elife, 4, 2015
|
|
7QS5
 
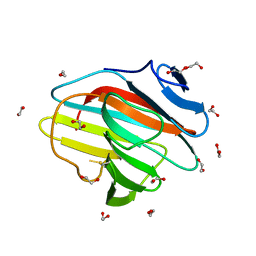 | | Crystal structure of B30.2 PRYSPRY domain of TRIM67 | | Descriptor: | 1,2-ETHANEDIOL, Tripartite motif-containing protein 67 | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of TRIM family PRYSPRY domains and its implications for E3-ligand design.
J Struct Biol X, 12, 2025
|
|
8CPJ
 
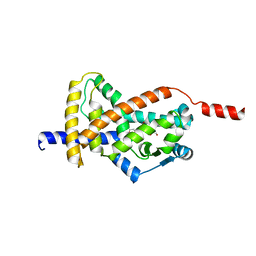 | | Crystal structure of PPAR gamma (PPARG) in an inactive form | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
1EJC
 
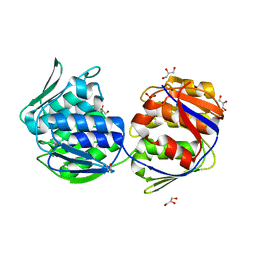 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | Authors: | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
1EVT
 
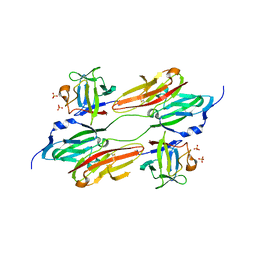 | | CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 1), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 1), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EWK
 
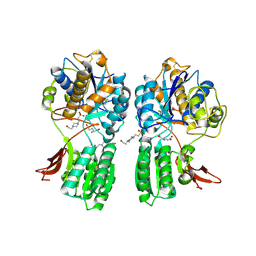 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
9FNV
 
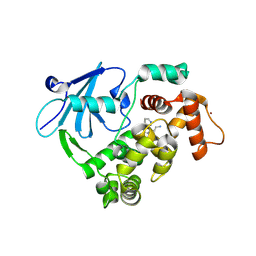 | |
8EM1
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA Unbound | | Descriptor: | 1,2-ETHANEDIOL, PaqCI, DNA Unbound | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
1EIF
 
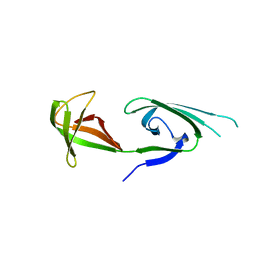 | | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A FROM METHANOCOCCUS JANNASCHII | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A | | Authors: | Kim, K.K, Hung, L.W, Yokota, H, Kim, R, Kim, S.H. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8C75
 
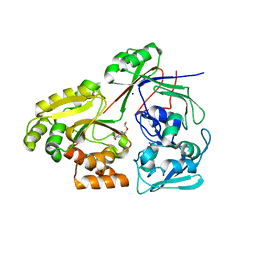 | | PBP AccA from A. tumefaciens Bo542 in apoform 3 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C6Y
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 2 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
