9QCW
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K51A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the Active Site of Class 3 L-Asparaginase by Mutagenesis: Mutations of the Ser-Lys Tandems of ReAV.
Biomolecules, 15, 2025
|
|
1EHI
 
 | | D-ALANINE:D-LACTATE LIGASE (LMDDL2) OF VANCOMYCIN-RESISTANT LEUCONOSTOC MESENTEROIDES | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE:D-LACTATE LIGASE, ... | | Authors: | Kuzin, A.P, Sun, T, Jorczak-Baillass, J, Healy, V.L, Walsh, C.T, Knox, J.R. | | Deposit date: | 2000-02-21 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Enzymes of vancomycin resistance: the structure of D-alanine-D-lactate ligase of naturally resistant Leuconostoc mesenteroides.
Structure Fold.Des., 8, 2000
|
|
9SL0
 
 | | Crystal structure of HLA-A0201 in complex with peptide LLWNGPMAV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Liu, Y, Gfeller, D, Guillaume, P, Larabi, A, Lau, K, Pojer, F. | | Deposit date: | 2025-09-02 | | Release date: | 2025-09-17 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HLA-A0201 in complex with peptide LLWNGPMAV
To Be Published
|
|
9QPZ
 
 | | KRAS-WT(1-169) - GDP IN COMPLEX WITH compound (R)-1 | | Descriptor: | (4~{R})-4-[[(1~{S},5~{R})-3-[7-(8-ethynyl-7-fluoranyl-3-oxidanyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]carbonyl]-3,3-dimethyl-oxetan-2-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Ostermann, N. | | Deposit date: | 2025-03-31 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.312 Å) | | Cite: | Promise and Challenge of beta-Lactone Electrophiles to Target Aspartate 12 of Mutant KRAS G12D .
J.Med.Chem., 68, 2025
|
|
9QEY
 
 | | Cryo-EM structure of the actin filament bound by a single Coronin-1B molecule. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-11 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
9S6B
 
 | | Aeropyrum pernix acylaminoacyl peptidase co-crystallized with meropenem. | | Descriptor: | (2R,2'R)-3,3'-oxydipropane-1,2-diol, Acylamino-acid-releasing enzyme | | Authors: | Harmat, V, Takacs, L, Kiss-Szeman, A, Banoczi, Z, Jakli, I, Hosogi, N, Traore, D.A.K, Perczel, A, Menyhard, D.K. | | Deposit date: | 2025-07-31 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding Pro-miscuity of acylpeptide hydrolase, structural analysis of a detoxifying serine hydrolase.
Protein Sci., 34, 2025
|
|
9PPV
 
 | | In situ MicroED structure of human eosinophil major basic protein-1 | | Descriptor: | Bone marrow proteoglycan, CHLORIDE ION | | Authors: | Yang, J.E, Bingman, C.A, Mitchell, J, Mosher, D, Wright, E.R. | | Deposit date: | 2025-07-22 | | Release date: | 2025-09-03 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | In situ MicroED structure of the human eosinophil major basic protein-1
To Be Published
|
|
9R1P
 
 | | Crystal structure of STUB1 complexed with a compound molecule | | Descriptor: | 1-[3-[1-methyl-4-(pyridin-3-ylmethylamino)pyrazolo[3,4-d]pyrimidin-6-yl]phenyl]-3-[3-(trifluoromethyl)phenyl]thiourea, E3 ubiquitin-protein ligase CHIP | | Authors: | Chu, Y, Jiang, H. | | Deposit date: | 2025-04-28 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A rapid imaging-based screen for induced-proximity degraders identifies a potent degrader of oncoprotein SKP2.
Nat.Biotechnol., 2025
|
|
9QF1
 
 | | Structure of CHIP E3 ubiquitin ligase TPR domain in complex with compound 2 | | Descriptor: | E3 ubiquitin-protein ligase CHIP, SULFATE ION, ~{N}-methyl-~{N}-[(1~{R})-1-pyridin-2-yl-3-pyrrolidin-1-yl-propyl]-5-[4,5,6,7-tetrakis(fluoranyl)-1~{H}-indol-3-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Breed, J. | | Deposit date: | 2025-03-11 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Discovery of Small-Molecule Ligands for the E3 Ligase STUB1/CHIP from a DNA-Encoded Library Screen.
Acs Med.Chem.Lett., 16, 2025
|
|
9O0J
 
 | | HtaA CR1 domain from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ford, J, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2025-04-02 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of heme scavenging by the ChtA and HtaA hemophores in Corynebacterium diphtheriae.
J.Biol.Chem., 301, 2025
|
|
1EYQ
 
 | | Chalcone isomerase and naringenin | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1, NARINGENIN, SULFATE ION | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
8SZS
 
 | | Cat DHX9 bound to GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Development of assays to support identification and characterization of modulators of DExH-box helicase DHX9.
Slas Discov, 28, 2023
|
|
1EVL
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH A THREONYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1ELD
 
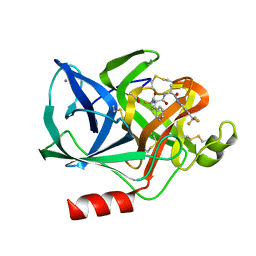 | | Structural analysis of the active site of porcine pancreatic elastase based on the x-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors | | Descriptor: | ACETIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
1EJM
 
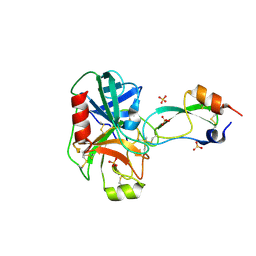 | | CRYSTAL STRUCTURE OF THE BPTI ALA16LEU MUTANT IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | BETA-TRYPSIN, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Otlewski, J, Smalas, A, Helland, R, Grzesiak, A, Krowarsch, D. | | Deposit date: | 2000-03-03 | | Release date: | 2001-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substitutions at the P(1) position in BPTI strongly affect the association energy with serine proteinases.
J.Mol.Biol., 301, 2000
|
|
1ELS
 
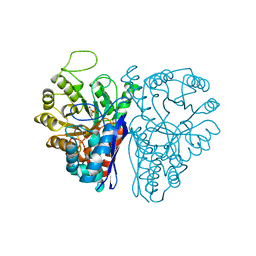 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
8YQ3
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | Authors: | Qiang, G, Zheng, Y.C, Feng, L, Yu, H.L. | | Deposit date: | 2024-03-19 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Loop Engineering Facilitates the Nicotinamide Cofactors Utilization of BVMOs
To Be Published
|
|
8VS7
 
 | | Crystal structure of ADI-19425 Fab in complex with anti-idiotypic 2C1 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2C1 Heavy Chain, 2C1 Light Chain, ... | | Authors: | Kher, G, Homad, L.J, McGuire, A.T, Pancera, M. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A novel RSV vaccine candidate derived from anti-idiotypic antibodies targets putative neutralizing B-cells in bulk PBMCs
To Be Published
|
|
1EVQ
 
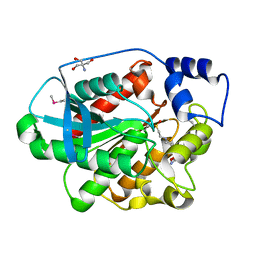 | | THE CRYSTAL STRUCTURE OF THE THERMOPHILIC CARBOXYLESTERASE EST2 FROM ALICYCLOBACILLUS ACIDOCALDARIUS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SERINE HYDROLASE | | Authors: | De Simone, G, Galdiero, S, Manco, G, Lang, D, Rossi, M, Pedone, C. | | Deposit date: | 2000-04-20 | | Release date: | 2000-11-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase.
J.Mol.Biol., 303, 2000
|
|
1EYP
 
 | | CHALCONE ISOMERASE | | Descriptor: | CHALCONE-FLAVONONE ISOMERASE 1 | | Authors: | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-05-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
7M73
 
 | |
8YU5
 
 | | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of non-activated thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q)
To Be Published
|
|
8C6U
 
 | | PBP AccA-G145YG440Q mutant from A. tumefaciens Bo542 in apoform 4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
1ECN
 
 | |
8FB4
 
 | | Structure of an alternating AT 16-mer bound by diamidine DB1476: 5'-GCTGGATATATCCAGC-3 | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), CALCIUM ION, DNA (5'-D(*GP*CP*TP*GP*GP*AP*TP*AP*TP*AP*TP*CP*CP*AP*GP*C)-3') | | Authors: | Terrell, J.R, Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
