2JUY
 
 | | NMR ensemble of Neopetrosiamide A | | Descriptor: | Neopetrosiamide A | | Authors: | Austin, P, Williams, D.E, Heller, M, McIntosh, L.P, Andersen, R.J, Roberge, M, Roskelley, C.D. | | Deposit date: | 2007-09-05 | | Release date: | 2008-08-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR ensemble of Neopetrosiamide A
To be Published
|
|
2JUZ
 
 | | Solution NMR structure of HI0947 from Haemophilus influenzae, Northeast Structural Genomics Consortium Target IR123 | | Descriptor: | UPF0352 protein HI0840 | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-09 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of HI0947 from Haemophilus influenzae.
To be Published
|
|
2JV0
 
 | | SET domain of RIZ1 tumor suppressor (PRDM2) | | Descriptor: | PR domain zinc finger protein 2 | | Authors: | Briknarova, K. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-22 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the SET domain from RIZ1 tumor suppressor
Biochem.Biophys.Res.Commun., 366, 2008
|
|
2JV1
 
 | |
2JV2
 
 | |
2JV3
 
 | |
2JV4
 
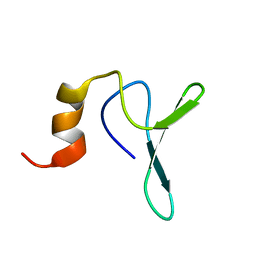 | | Structure Characterisation of PINA WW Domain and Comparison with other Group IV WW Domains, PIN1 and ESS1 | | Descriptor: | Peptidyl-prolyl cis/trans isomerase | | Authors: | Ng, C.A, Kato, Y, Tanokura, M, Brownlee, R.T.C. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of PinA WW domain and a comparison with other Group IV WW domains, Pin1 and Ess1
Biochim.Biophys.Acta, 1784, 2008
|
|
2JV5
 
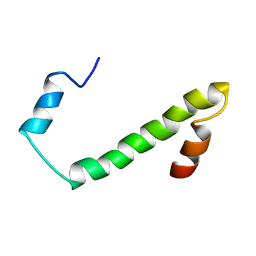 | | Nogo54 | | Descriptor: | Reticulon-4 | | Authors: | Li, M, Song, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of buffer-soluble Nogo54: a molecular mimic of Nogo-66 to inhibit CNS neuron regeneration
To be Published
|
|
2JV6
 
 | | YF ED3 Protein NMR Structure | | Descriptor: | Envelope protein E | | Authors: | Volk, D.E, Gandham, S.H.A, May, F.J, Anderson, A, Barrett, A.D.T, Gorenstein, D.G. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Yellow Fever Virus Envelope Protein Domain III: A Convergence of Structure and Phylogenetics
To be Published
|
|
2JV7
 
 | |
2JV8
 
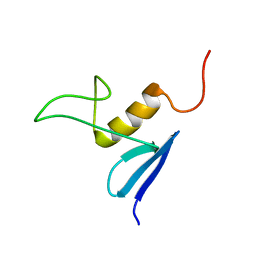 | | Solution structure of protein NE1242 from Nitrosomonas europaea. Northeast Structural Genomics Consortium Target NeT4 | | Descriptor: | Uncharacterized protein NE1242 | | Authors: | Wu, Y, Yee, A, Zeri, A.C, Guido, V, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein NE1242 from Nitrosomonas europaea.
To be Published
|
|
2JV9
 
 | |
2JVA
 
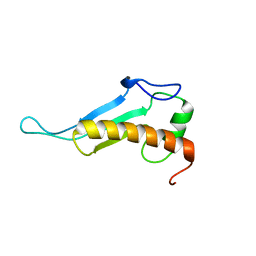 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | Descriptor: | Peptidyl-tRNA hydrolase domain protein | | Authors: | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
2JVB
 
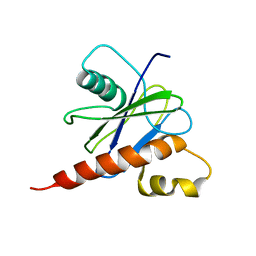 | |
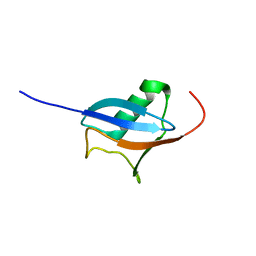
2JVC
 
 | |
2JVD
 
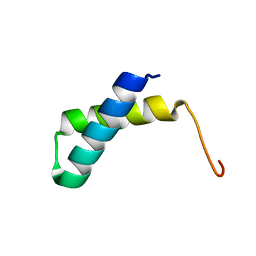 | | Solution NMR structure of the folded N-terminal fragment of UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384-1-46 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Sharma, S, Huang, Y.J, Zhao, L, Owens, L.A, Stokes, K, Jiang, M, Xiao, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis.
Proteins, 72, 2008
|
|
2JVE
 
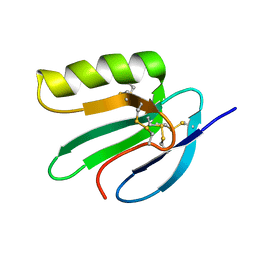 | | Solution structure of the extracellular domain of Prod1, a protein implicated in proximodistal identity during amphibian limb regeneration | | Descriptor: | Prod 1 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C. | | Deposit date: | 2007-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phylogenetics of Prod1, a member of the three-finger protein superfamily implicated in salamander limb regeneration.
Plos One, 4, 2009
|
|
2JVF
 
 | | Solution structure of M7, a computationally-designed artificial protein | | Descriptor: | de novo protein M7 | | Authors: | Stordeur, C, Dalluege, R, Birkenmeier, O, Wienk, H, Rudolph, R, Lange, C, Luecke, C. | | Deposit date: | 2007-09-19 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the artificial protein M7 matches the computationally designed model
Proteins, 72, 2008
|
|
2JVG
 
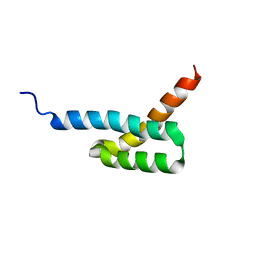 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2JVH
 
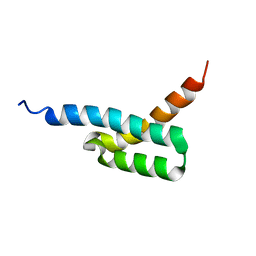 | | Structure of C3-binding domain 4 of S. aureus protein Sbi | | Descriptor: | IgG-binding protein SBI | | Authors: | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2JVI
 
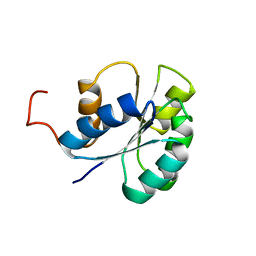 | |
2JVJ
 
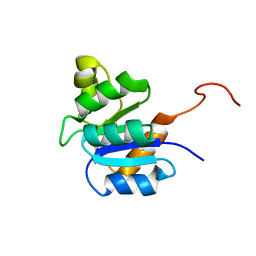 | |
2JVK
 
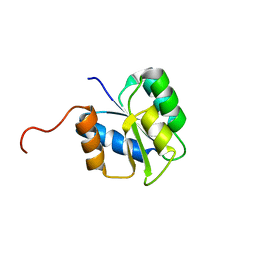 | |
2JVL
 
 | | NMR structure of the C-terminal domain of MBF1 of Trichoderma reesei | | Descriptor: | TrMBF1 | | Authors: | Kopke Salinas, R, Tomaselli, S, Camilo, C.M, Valencia, E.Y, Farah, C.S, El-Dorry, H, Chambergo, F.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of multiprotein bridging factor 1 (MBF1) of Trichoderma reesei.
Proteins, 75, 2009
|
|
2JVM
 
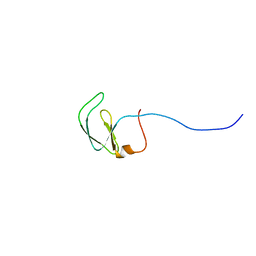 | | Solution NMR structure of Rhodobacter sphaeroides protein RHOS4_26430. Northeast Structural Genomics Consortium target RhR95 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Sukumaran, D, Zhang, Q, Parish, D, Xu, D, Wang, H, Janjua, H, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Rhodobacter sphaeroides protein RHOS4_26430.
To be Published
|
|
