7Q4G
 
 | | Structure of coproheme decarboxylase from Corynebacterium dipththeriae Y135A mutant in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Coproheme decarboxylase from Corynebacterium diphtheriae Y135A mutant in complex with coproheme, DI(HYDROXYETHYL)ETHER | | Authors: | Michlits, H, Valente, N, Mlynek, G, Hofbauer, S. | | Deposit date: | 2021-10-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Initial Steps to Engineer Coproheme Decarboxylase to Obtain Stereospecific Monovinyl, Monopropionyl Deuterohemes.
Front Bioeng Biotechnol, 9, 2021
|
|
6BOK
 
 | |
7Q8Y
 
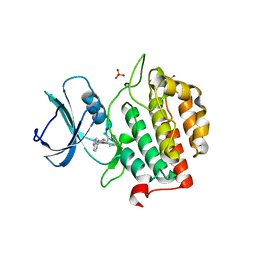 | | Crystal structure of TTBK2 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7Q8Z
 
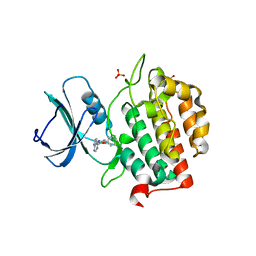 | | Crystal structure of TTBK2 in complex with VNG1.33 (compound 27) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-(4-phenoxyphenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7Q90
 
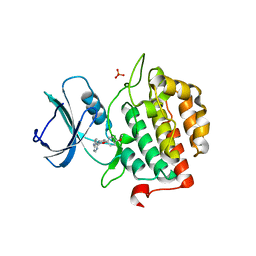 | | Crystal structure of TTBK2 in complex with VNG1.63 (compound 32) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 2, ~{N}-[4-(4-methoxyphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
6C9X
 
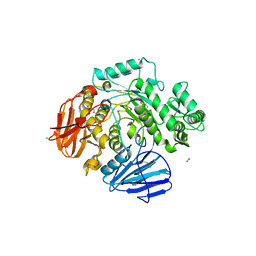 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | Descriptor: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6C69
 
 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6E
 
 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
4P93
 
 | | Structure of Dienelactone Hydrolase at 1.85 A resolution crystallised in the C2 space group | | Descriptor: | Carboxymethylenebutenolidase | | Authors: | Porter, J.L, Carr, P.D, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallization of dienelactone hydrolase in two space groups: structural changes caused by crystal packing.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7URP
 
 | |
6OQA
 
 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4PKS
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6BQT
 
 | |
1KS0
 
 | | The First Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | Matrix Metalloproteinase 2 | | Authors: | Gehrmann, M, Briknarova, K, Banyai, L, Patthy, L, Llinas, M. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The col-1 module of human matrix metalloproteinase-2 (MMP-2): structural/functional relatedness between gelatin-binding fibronectin type II modules and lysine-binding kringle domains.
Biol.Chem., 383, 2002
|
|
4PLH
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with oxamate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, OXAMIC ACID, SODIUM ION, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PK3
 
 | | tubulin acetyltransferase complex with bisubstrate analog | | Descriptor: | ACETYL-SER-ASP-(N-ACETYL-LYS)-THR-NH2 PEPTIDE, Alpha-tubulin N-acetyltransferase 1, COENZYME A | | Authors: | Szyk, A, Roll-Mecak, A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase.
Cell, 157, 2014
|
|
7UWU
 
 | | Starch adherence system protein 6 (Sas6) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Starch Adherence System protein 6 (Sas6), ... | | Authors: | Photenhauer, A.L, Koropatkin, N.M. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Ruminococcus bromii amylosome protein Sas6 binds single and double helical alpha-glucan structures in starch.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3W34
 
 | | Ternary complex of Thermus thermophilus HB8 uridine-cytidine kinase with substrates | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Biochemical Studies on the Reaction Mechanism of Uridine-Cytidine Kinase
Protein J., 34, 2015
|
|
4P5S
 
 | | Structure of reduced W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
6C18
 
 | | FGFR1 kinase complex with inhibitor SN37115 | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-7-[(propan-2-yl)amino]-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
6C63
 
 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.900028 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
8BYJ
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8GV1
 
 | | Crystal structure of anti-FX IgG fab with FAST-Ig mutations | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Anti-factor X IgG fab heavy chain, ... | | Authors: | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | Deposit date: | 2022-09-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.186 Å) | | Cite: | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
4PFC
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-(5-{5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}-2-fluorophenyl)-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
