7M06
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, Y110F mutant with R,R-bislysine bound at the allosteric site at 2.7 Angstrom | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Sanders, D.A.R. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | B-FACTOR ANALYSIS SUGGEST THAT L-LYSINE AND R, R-BISLYSINE ALLOSTERICALLY INHIBIT Cj.DHDPS ENZYME BY DECREASING PROTEIN DYNAMICS
To Be Published
|
|
5VQ8
 
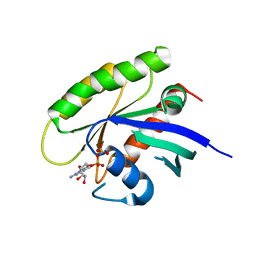 | | Crystal structure of human WT-KRAS in complex with GDP (EDTA soaked) | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Xu, S, Long, B, Boris, G, Ni, S, Kennedy, M.A. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8W1J
 
 | | Crystal Structure of a fatty acid decarboxylase from Corynebacterium lipophiloflavum in complex with palmitic acid | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fatty acid decarboxylase, ... | | Authors: | Generoso, W.C, Miyamoto, R.Y, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2024-02-16 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coordinated conformational changes in P450 decarboxylases enable hydrocarbons production from renewable feedstocks.
Nat Commun, 16, 2025
|
|
6VV8
 
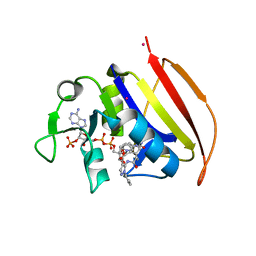 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB285 | | Descriptor: | 5-{[3-(1H-indol-3-yl)propanoyl]amino}-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
8VGB
 
 | |
8VYD
 
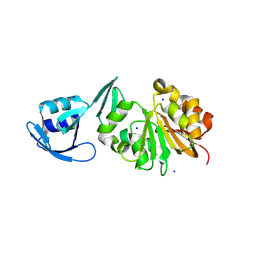 | | A novel synthase generates m4(2)C to stabilize the archaeal ribosome | | Descriptor: | SAM-dependent methyltransferase, UPF0020 family, SODIUM ION | | Authors: | Ho, P.S, Santangelo, T.J, Fluke, K. | | Deposit date: | 2024-02-08 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A novel N 4, N 4-dimethylcytidine in the archaeal ribosome enhances hyperthermophily.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6VVF
 
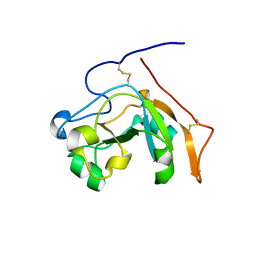 | |
5VQW
 
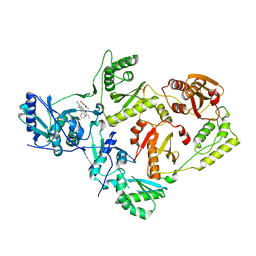 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acrylamide (JLJ685), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)prop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Kudalkar, S.N, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6VVT
 
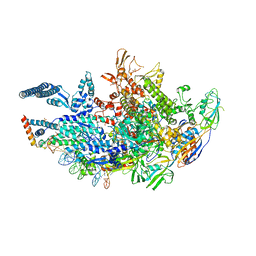 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8VIV
 
 | |
5INC
 
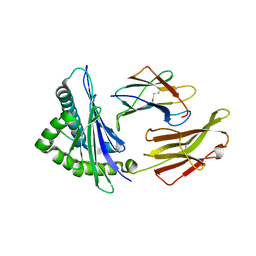 | | Crystal structure of HLA-B5801, a protective HLA allele for HIV-1 infection | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, HLA class I histocompatibility antigen, ... | | Authors: | Li, X, Wang, J.-H. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Crystal structure of HLA-B*5801, a protective HLA allele for HIV-1 infection.
Protein Cell, 7, 2016
|
|
7LUG
 
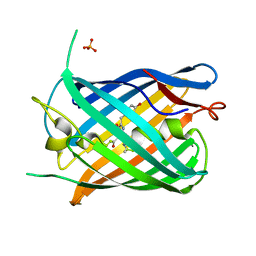 | | Crystal structure of the pnRFP B30Y mutant | | Descriptor: | PHOSPHATE ION, Red Fluorescent pnRFP B30Y mutant | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
8W1K
 
 | | Crystal Structure of a fatty acid decarboxylase from Corynebacterium lipophiloflavum in complex with oleic acid | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fatty acid decarboxylase, ... | | Authors: | Generoso, W.C, Miyamoto, R.Y, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2024-02-16 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coordinated conformational changes in P450 decarboxylases enable hydrocarbons production from renewable feedstocks.
Nat Commun, 16, 2025
|
|
8VBG
 
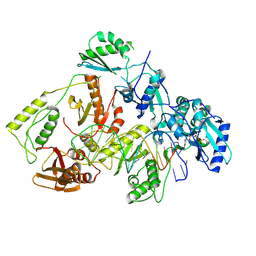 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
5INK
 
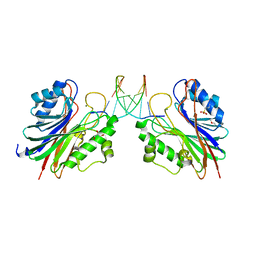 | |
8W1U
 
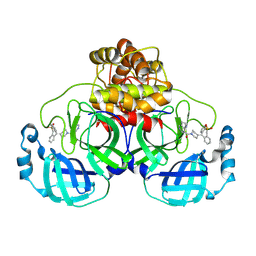 | | SARS-CoV-2 Main protease bound to non-covalent lead molecule NZ-804 | | Descriptor: | 11-[1-(1H-pyrrolo[3,2-c]pyridine-7-carbonyl)piperidin-4-ylidene]-6,11-dihydro-5H-5lambda~6~-dibenzo[b,e]thiepine-5,5-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
5IJE
 
 | | Crystal structure of Equine Serum Albumin in the presence of 30 mM zinc at pH 7.4 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Szlachta, K, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
8W1T
 
 | | SARS-CoV-2 Main protease bound to a non-covalent non-peptidic HTS hit | | Descriptor: | (5-bromopyridin-3-yl){4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, (5-bromopyridin-3-yl){4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}methanone, 1,2-ETHANEDIOL, ... | | Authors: | Bian, X, Tang, S, Sacchettini, J.C. | | Deposit date: | 2024-02-18 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An oral non-covalent non-peptidic inhibitor of SARS-CoV-2 Mpro ameliorates viral replication and pathogenesis in vivo.
Cell Rep, 43, 2024
|
|
5VD9
 
 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-097, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1R)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
2XZW
 
 | | STRUCTURE OF PII FROM SYNECHOCOCCUS ELONGATUS IN COMPLEX WITH 2- OXOGLUTARATE AT LOW 2-OG CONCENTRATIONS | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zeth, K, Chellamuthu, V.-R, Forchhammer, K, Fokina, O. | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of 2-Oxoglutarate Signaling by the Synechococcus Elongatus Pii Signal Transduction Protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8VV8
 
 | | Crystal Structure of EHMT2 bound to EZM8266 | | Descriptor: | (2R)-1-(azetidin-1-yl)-3-(2-methoxy-5-{[4-methyl-6-(methylamino)pyrimidin-2-yl]amino}phenoxy)propan-2-ol, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Cocozaki, A, Farrow, N.A. | | Deposit date: | 2024-01-30 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combining EHMT and PARP Inhibition: A Strategy to Diminish Therapy-Resistant Ovarian Cancer Tumor Growth while Stimulating Immune Activation.
Mol.Cancer Ther., 23, 2024
|
|
5VDT
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
5IOY
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-(cyclopentylmethyl)pyrrolidine-1-carboxamide at 1.77A resolution | | Descriptor: | N-(cyclopentylmethyl)pyrrolidine-1-carboxamide, SULFATE ION, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
8VBD
 
 | | Kinetic intermediate states of HIV-1 RT DNA synthesis captured by cryo-EM | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (38-MER), HIV-1 reverse transcriptase P51 subunit, ... | | Authors: | Vergara, S, Zhou, X, Santiago, U, Conway, J.F, Sluis-Cremer, N, Calero, G. | | Deposit date: | 2023-12-12 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of deoxynucleotide addition by HIV-1 RT during reverse transcription.
Nat Commun, 15, 2024
|
|
7LQO
 
 | | Crystal structure of a genetically encoded red fluorescent peroxynitrite biosensor, pnRFP | | Descriptor: | PHOSPHATE ION, red fluorescent peroxynitrite biosensor pnRFP | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-14 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
