6NMT
 
 | | Non-Blocking Fab 3 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 3 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 3 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NMS
 
 | | Blocking Fab 136 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 136 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 136 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6SZ6
 
 | | Chaetomium thermophilum beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Mohsin, I, Poudel, N, Papageorgiou, A.C. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Crystal Structure of a GH3 beta-Glucosidase from the Thermophilic Fungus Chaetomium thermophilum .
Int J Mol Sci, 20, 2019
|
|
6XP7
 
 | |
6XPU
 
 | |
6OEI
 
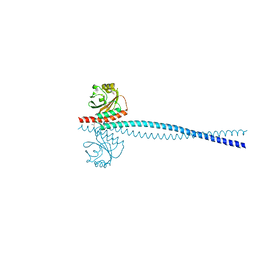 | | Yeast Spc42 N-terminal coiled-coil fused to PDB: 3K2N | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Spindle pole body component SPC42,Sigma-54-dependent transcriptional regulator | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
8V1X
 
 | | Crystal structure of polyketide synthase (PKS) thioreductase domain from Streptomyces coelicolor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pereira, J.H, Dan, Q, Keasling, J, Adams, P.D. | | Deposit date: | 2023-11-21 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A polyketide-based biosynthetic platform for diols, amino alcohols and hydroxy acids
Nat Catal, 8, 2025
|
|
6XP4
 
 | |
6XPS
 
 | |
6XPT
 
 | |
7YGG
 
 | |
5K9H
 
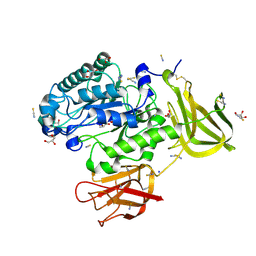 | | Crystal structure of a glycoside hydrolase 29 family member from an unknown rumen bacterium | | Descriptor: | 0940_GH29, GLYCEROL, SODIUM ION, ... | | Authors: | Summers, E.L, Arcus, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8DFZ
 
 | |
6I0I
 
 | |
6OBD
 
 | |
7UI4
 
 | |
7R97
 
 | | Crystal structure of postcleavge complex of Escherichia coli RNase III | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Dharavath, S, Shaw, G.X, Ji, X. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg 2+ -ion catalysis.
Rna Biol., 19, 2022
|
|
6NMV
 
 | | Non-Blocking Fab 218 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 218 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 218 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NV6
 
 | |
7RHO
 
 | | Human IgG1 Fc fragment, hinge-free, expressed in E. coli | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fc fragment of human IgG1 | | Authors: | Gallagher, D.T. | | Deposit date: | 2021-07-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of glycans and hinge on dynamics in the IgG1 Fc.
J.Biomol.Struct.Dyn., 2023
|
|
6Y9S
 
 | | Crystal structure of GSK-3b in complex with the imidazo[1,5-a]pyridine-3-carboxamide inhibitor 16 | | Descriptor: | ACETATE ION, Glycogen synthase kinase-3 beta, ~{N}-(oxan-4-ylmethyl)-6-(5-propan-2-yloxypyridin-3-yl)imidazo[1,5-a]pyridine-3-carboxamide | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
2IBL
 
 | |
6NMU
 
 | | Kick-Off Fab 115 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 115 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 115 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
6NXV
 
 | | Crystal structure of the theta class glutathione S-transferase from the citrus canker pathogen Xanthomonas axonopodis pv. citri, apo form | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Glutathione S-transferase | | Authors: | Hilario, E, De Keyser, S, Fan, L. | | Deposit date: | 2019-02-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical characterization of a glutathione transferase from the citrus canker pathogen Xanthomonas.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8I5O
 
 | |
