6K1N
 
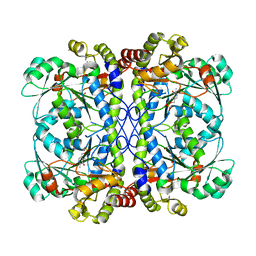 | | PLP-bound form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
3NQD
 
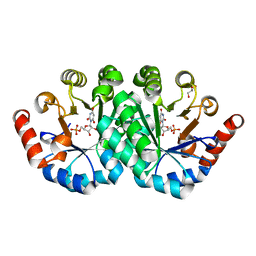 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3NQC
 
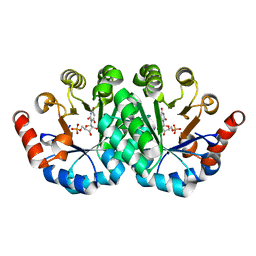 | | Crystal structure of the mutant I96S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
4ADX
 
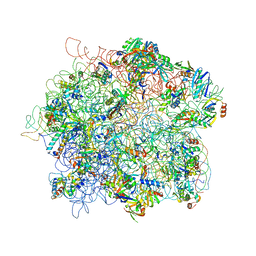 | | The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 | | Descriptor: | 23S Ribosomal RNA EXPANSION SEGMENTS, 23S ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Godinic-Mikulcic, V, Crnkovic, A, Ibba, M, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-Em Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 and Implications for Ribosome Evolution
J.Mol.Biol., 418, 2012
|
|
7EIM
 
 | |
7EJT
 
 | |
7EJP
 
 | |
7EKX
 
 | |
7EKW
 
 | |
3KGC
 
 | | Isolated ligand binding domain dimer of GluA2 ionotropic glutamate receptor in complex with glutamate, LY 404187 and ZK 200775 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-[(2S)-2-(4'-cyanobiphenyl-4-yl)propyl]propane-2-sulfonamide, ... | | Authors: | Sobolevsky, A.I, Rosconi, M.P, Gouaux, E. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor
Nature, 462, 2009
|
|
7EDK
 
 | |
3W07
 
 | |
3WV7
 
 | | HcgE from Methanothermobacter marburgensis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hmd co-occurring protein HcgE | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WV8
 
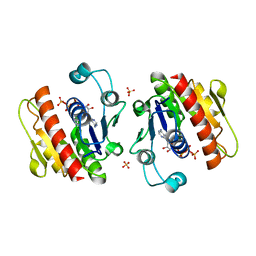 | | ATP-bound HcgE from Methanothermobacter marburgensis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hmd co-occurring protein HcgE, SULFATE ION | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WV9
 
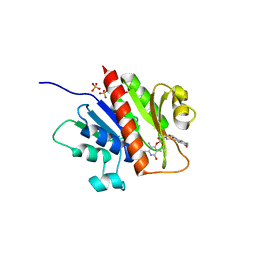 | | Guanylylpyridinol (GP)- and ATP-bound HcgE from Methanothermobacter marburgensis | | Descriptor: | 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, ADENOSINE-5'-TRIPHOSPHATE, Hmd co-occurring protein HcgE, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
2MOA
 
 | |
7E0L
 
 | |
2LR9
 
 | |
2YYF
 
 | | Purification and structural characterization of a D-amino acid containing conopeptide, marmophine, from Conus marmoreus | | Descriptor: | M-conotoxin mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C. | | Deposit date: | 2007-04-29 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a D-amino acid-containing conopeptide, conomarphin, from Conus marmoreus.
Febs J., 275, 2008
|
|
1PU1
 
 | | Solution structure of the Hypothetical protein mth677 from Methanothermobacter Thermautotrophicus | | Descriptor: | Hypothetical protein MTH677 | | Authors: | Blanco, F.J, Yee, A, Campos-Olivas, R, Devos, D, Valencia, A, Arrowsmith, C.H, Rico, M. | | Deposit date: | 2003-06-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein Mth677 from Methanobacterium thermoautotrophicum: A novel {alpha}+{beta} fold
Protein Sci., 13, 2004
|
|
2EVU
 
 | | Crystal structure of aquaporin AqpM at 2.3A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-10-31 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2F2B
 
 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1OQK
 
 | | Structure of Mth11: A homologue of human RNase P protein Rpp29 | | Descriptor: | conserved protein MTH11 | | Authors: | Boomershine, W.P, McElroy, C.A, Tsai, H, Gopalan, V, Foster, M.P. | | Deposit date: | 2003-03-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1TR8
 
 | |
3FPE
 
 | | Crystal Structure of MtNAS in complex with thermonicotianamine | | Descriptor: | BROMIDE ION, N-[(3S)-3-{[(3S)-3-amino-3-carboxypropyl]amino}-3-carboxypropyl]-L-glutamic acid, Putative uncharacterized protein | | Authors: | Dreyfus, C, Pignol, D, Arnoux, P. | | Deposit date: | 2009-01-05 | | Release date: | 2009-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
