1PTG
 
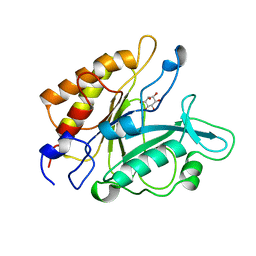 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C IN COMPLEX WITH MYO-INOSITOL | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W, Ryan, M, Bullock, T.L, Griffith, O.H. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol.
EMBO J., 14, 1995
|
|
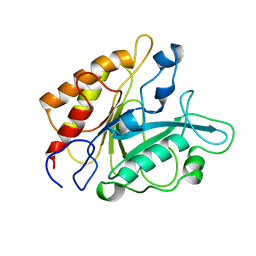
1PTD
 
 | |
1PSJ
 
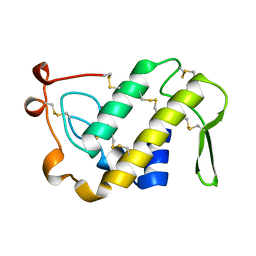 | | ACIDIC PHOSPHOLIPASE A2 FROM AGKISTRODON HALYS PALLAS | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Wang, X.Q, Lin, Z.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acidic phospholipase A2 from the venom of Agkistrodon halys pallas at 2.0 A resolution.
J.Mol.Biol., 255, 1996
|
|
1GAC
 
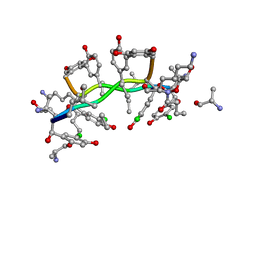 | | NMR structure of asymmetric homodimer of a82846b, a glycopeptide antibiotic, complexed with its cell wall pentapeptide fragment | | Descriptor: | CELL WALL PENTAPEPTIDE, CHLOROORIENTICIN A, vancosamine, ... | | Authors: | Kline, A.D, Prowse, W.G, Skelton, M.A, Loncharich, R.J. | | Deposit date: | 1995-05-24 | | Release date: | 1996-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformation of A82846B, a Glycopeptide Antibiotic, Complexed with its Cell Wall Fragment: An Asymmetric Homodimer Determined Using NMR Spectroscopy.
Biochemistry, 34, 1995
|
|
1ESF
 
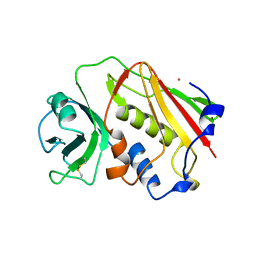 | | STAPHYLOCOCCAL ENTEROTOXIN A | | Descriptor: | CADMIUM ION, STAPHYLOCOCCAL ENTEROTOXIN A | | Authors: | Schad, E.M, Svensson, L.A. | | Deposit date: | 1995-05-25 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superantigen staphylococcal enterotoxin type A.
EMBO J., 14, 1995
|
|
1GEC
 
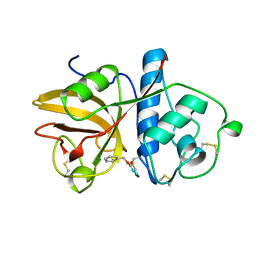 | | GLYCYL ENDOPEPTIDASE-COMPLEX WITH BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE COVALENTLY BOUND TO CYSTEINE 25 | | Descriptor: | BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE INHIBITOR, GLYCYL ENDOPEPTIDASE | | Authors: | Ohara, B.P, Hemmings, A.M, Buttle, D.J, Pearl, L.H. | | Deposit date: | 1995-05-25 | | Release date: | 1995-12-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity.
Biochemistry, 34, 1995
|
|
4BJL
 
 | |
1SGP
 
 | |
1BCM
 
 | |
1HUE
 
 | | HISTONE-LIKE PROTEIN | | Descriptor: | HU PROTEIN | | Authors: | Vis, H, Mariani, M, Vorgias, C.E, Wilson, K.S, Kaptein, R, Boelens, R. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HU protein from Bacillus stearothermophilus.
J.Mol.Biol., 254, 1995
|
|
1SGQ
 
 | |
1BJM
 
 | |
3BJL
 
 | |
1BCO
 
 | | BACTERIOPHAGE MU TRANSPOSASE CORE DOMAIN | | Descriptor: | BACTERIOPHAGE MU TRANSPOSASE | | Authors: | Rice, P.A, Mizuuchi, K. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration.
Cell(Cambridge,Mass.), 82, 1995
|
|
1ATL
 
 | | Structural interaction of natural and synthetic inhibitors with the VENOM METALLOPROTEINASE, ATROLYSIN C (FORM-D) | | Descriptor: | CALCIUM ION, O-methyl-N-[(2S)-4-methyl-2-(sulfanylmethyl)pentanoyl]-L-tyrosine, Snake venom metalloproteinase atrolysin-D, ... | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1GYA
 
 | | N-GLYCAN AND POLYPEPTIDE NMR SOLUTION STRUCTURES OF THE ADHESION DOMAIN OF HUMAN CD2 | | Descriptor: | HUMAN CD2, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wyss, D.F, Choi, J.S, Wagner, G. | | Deposit date: | 1995-05-26 | | Release date: | 1996-11-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Conformation and function of the N-linked glycan in the adhesion domain of human CD2.
Science, 269, 1995
|
|
1SGR
 
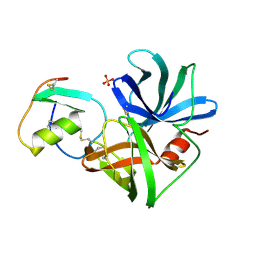 | |
1APF
 
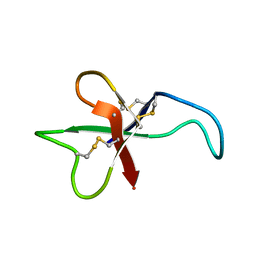 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
1HIJ
 
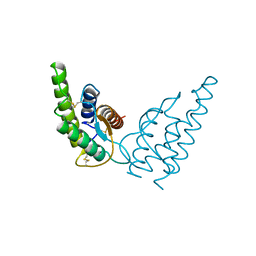 | |
1HXN
 
 | |
1CIY
 
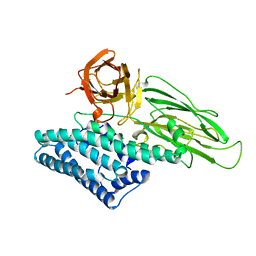 | |
1GPC
 
 | | CORE GP32, DNA-BINDING PROTEIN | | Descriptor: | PROTEIN (CORE GP32), ZINC ION | | Authors: | Shamoo, Y, Friedman, A.M, Parsons, M.R, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1995-06-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA.
Nature, 376, 1995
|
|
1HIK
 
 | | INTERLEUKIN-4 (WILD-TYPE) | | Descriptor: | INTERLEUKIN-4 | | Authors: | Mueller, T, Buehner, M. | | Deposit date: | 1995-06-01 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human interleukin-4 and variant R88Q: phasing X-ray diffraction data by molecular replacement using X-ray and nuclear magnetic resonance models.
J.Mol.Biol., 247, 1995
|
|
1ALI
 
 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1FPL
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE (D-FRUCTOSE-1,6-BISPHOSPHATE 1-PHOSPHOHYDROLASE) COMPLEXED WITH AMP, 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND THALLIUM IONS (10 MM) | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Lipscomb, W.N. | | Deposit date: | 1995-06-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
