2GGG
 
 | |
2GGH
 
 | |
2GGI
 
 | |
2GGJ
 
 | |
2GGK
 
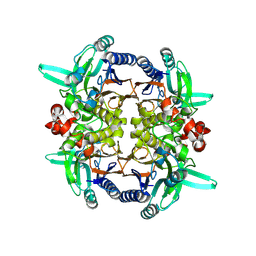 | |
2GGL
 
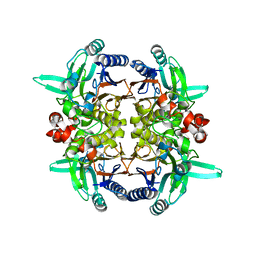 | |
2GGM
 
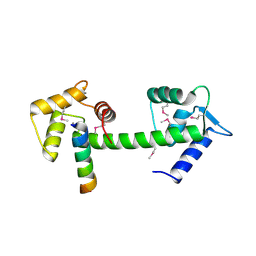 | | Human centrin 2 xeroderma pigmentosum group C protein complex | | Descriptor: | CALCIUM ION, Centrin-2, DNA-repair protein complementing XP-C cells | | Authors: | Thompson, J.R. | | Deposit date: | 2006-03-24 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the human centrin 2-xeroderma pigmentosum group C protein complex.
J.Biol.Chem., 281, 2006
|
|
2GGN
 
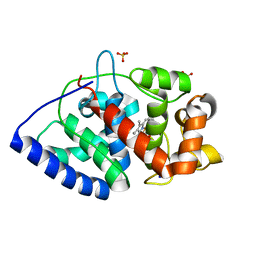 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
2GGO
 
 | | Crystal Structure of glucose-1-phosphate thymidylyltransferase from Sulfolobus tokodaii | | Descriptor: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of glucose-1-phosphate thymidylyltransferase from
Sulfolobus tokodaii
To be published
|
|
2GGP
 
 | | Solution structure of the Atx1-Cu(I)-Ccc2a complex | | Descriptor: | COPPER (I) ION, Metal homeostasis factor ATX1, Probable copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Cantini, F, Felli, I.C, Gonnelli, L, Hadjiliadis, N, Pierattelli, R, Rosato, A, Voulgaris, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-03-24 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction.
Nat.Chem.Biol., 2, 2006
|
|
2GGQ
 
 | | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii | | Descriptor: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase, IODIDE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii
To be published
|
|
2GGR
 
 | |
2GGS
 
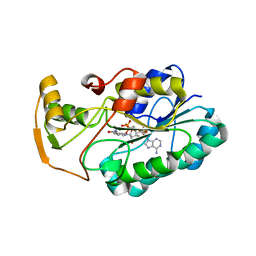 | | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii | | Descriptor: | 273aa long hypothetical dTDP-4-dehydrorhamnose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii
To be published
|
|
2GGT
 
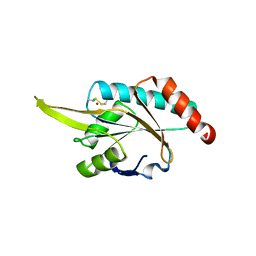 | | Crystal structure of human SCO1 complexed with nickel. | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SCO1 protein homolog, ... | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Martinelli, M, Palumaa, P, Wang, S. | | Deposit date: | 2006-03-24 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GGU
 
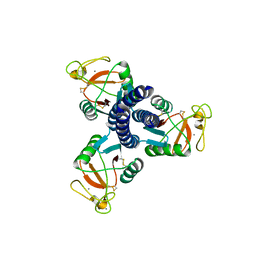 | |
2GGV
 
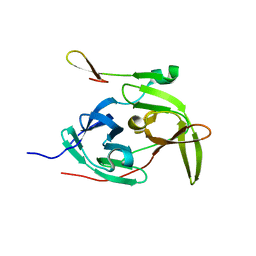 | | Crystal structure of the West Nile virus NS2B-NS3 protease, His51Ala mutant | | Descriptor: | non-structural protein 2B, non-structural protein 3 | | Authors: | Aleshin, A.E, Shiryaev, S.A, Strongin, A.Y, Liddington, R.C. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold.
Protein Sci., 16, 2007
|
|
2GGX
 
 | |
2GGZ
 
 | |
2GH0
 
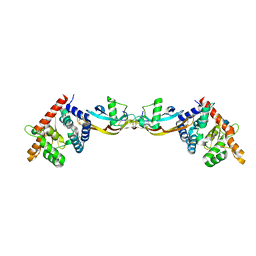 | | Growth factor/receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDNF family receptor alpha-3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
2GH1
 
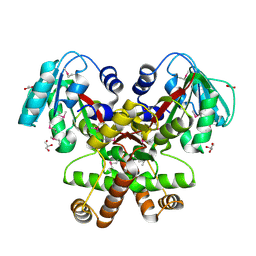 | | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20. | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase | | Authors: | Forouhar, F, Neely, H, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-24 | | Release date: | 2006-04-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20.
To be Published
|
|
2GH2
 
 | |
2GH4
 
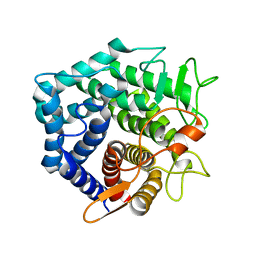 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
2GH5
 
 | | Crystal Structure of human Glutathione Reductase complexed with a Fluoro-Analogue of the Menadione Derivative M5 | | Descriptor: | 6-(3-METHYL-1,4-DIOXO-1,4-DIHYDRONAPHTHALEN-2-YL)HEXANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Winzer, A, Bauer, H, Schirmer, H, Davioud-Charvet, E. | | Deposit date: | 2006-03-25 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fluoro analogue of the menadione derivative 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme
J.Am.Chem.Soc., 128, 2006
|
|
2GH6
 
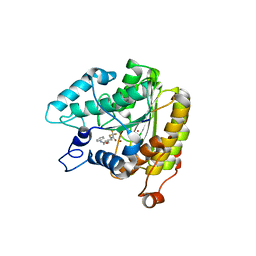 | | Crystal structure of a HDAC-like protein with 9,9,9-trifluoro-8-oxo-N-phenylnonan amide bound | | Descriptor: | 9,9,9-TRIFLUORO-8-OXO-N-PHENYLNONANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Riester, D, Wegener, D, Schwienhorst, A, Ficner, R. | | Deposit date: | 2006-03-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Complex structure of a bacterial class 2 histone deacetylase homologue with a trifluoromethylketone inhibitor.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2GH7
 
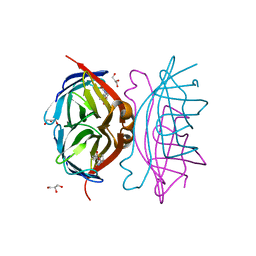 | | Epi-biotin complex with core streptavidin | | Descriptor: | BIOTIN, EPI-BIOTIN, GLYCEROL, ... | | Authors: | Le Trong, I, Aubert, D.G.L, Thomas, N.R, Stenkamp, R.E. | | Deposit date: | 2006-03-26 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The high-resolution structure of (+)-epi-biotin bound to streptavidin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
