6FYH
 
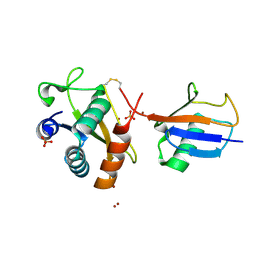 | | Disulfide between ubiquitin G76C and the E3 HECT ligase Huwe1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Jaeckl, M, Hartmann, M.D, Wiesner, S. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
J. Mol. Biol., 430, 2018
|
|
6FOQ
 
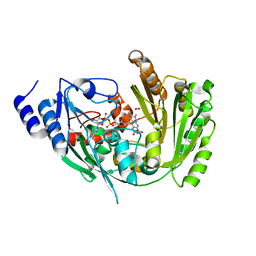 | |
6FNW
 
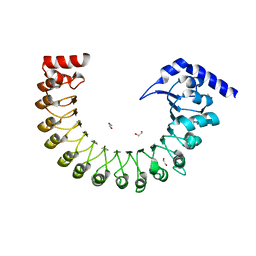 | | Structure of a volume-regulated anion channel of the LRRC8 family | | Descriptor: | 1,2-ETHANEDIOL, Volume-regulated anion channel subunit LRRC8A | | Authors: | Deneka, D, Sawicka, M, Lam, A.K.M, Paulino, C, Dutzler, R. | | Deposit date: | 2018-02-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a volume-regulated anion channel of the LRRC8 family.
Nature, 558, 2018
|
|
6FR5
 
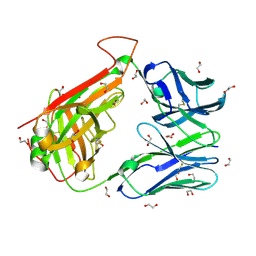 | | HA1.7 TCR Study of CDR Loop Flexibility | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | In Silicoand Structural Analyses Demonstrate That Intrinsic Protein Motions Guide T Cell Receptor Complementarity Determining Region Loop Flexibility.
Front Immunol, 9, 2018
|
|
6FS3
 
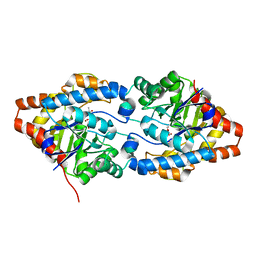 | | Phosphotriesterase PTE_A53_1 | | Descriptor: | (2~{S})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Bacterail Phosphotriesterase variant with high catalytic activity towards organophosphate nerve agents developed by use of structure-based design and molecular evolution
To Be Published
|
|
6FU8
 
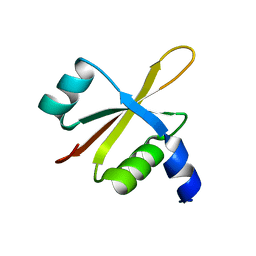 | |
6FUM
 
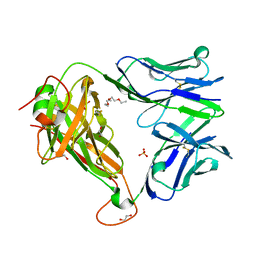 | |
6FVO
 
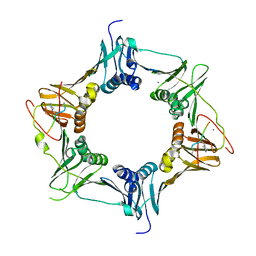 | | Mutant DNA polymerase sliding clamp from Mycobacterium tuberculosis with bound P7 peptide | | Descriptor: | Beta sliding clamp, CALCIUM ION, P7 peptide | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FUO
 
 | |
6FX2
 
 | | crystal structure of Pholiota squarrosa lectin in complex with a decasaccharide | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, lectin | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8HEA
 
 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
6FYC
 
 | |
6G01
 
 | |
6FOW
 
 | |
8HGI
 
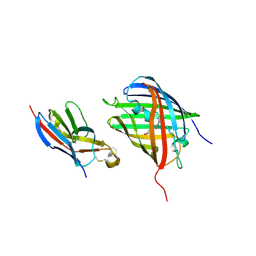 | | Crystal structure of VNAR aGFP14 in complex with GFP | | Descriptor: | GFP, VNAR aGFP14 | | Authors: | Zheng, P, Zhu, C, Jin, T. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selection, identification and crystal structure of shark-derived single-domain antibodies against a green fluorescent protein.
Int.J.Biol.Macromol., 247, 2023
|
|
6FRA
 
 | |
6G7L
 
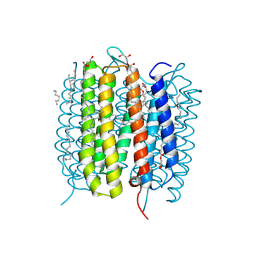 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 8.3 ms state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7H
 
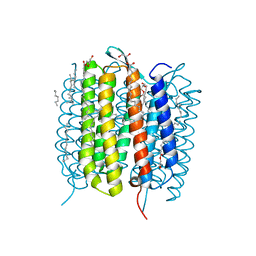 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: resting state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
5QDB
 
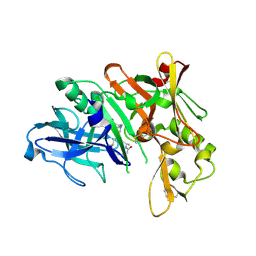 | | Crystal structure of BACE complex with BMC002 | | Descriptor: | (3S,14R,16S)-16-[(1R)-1-hydroxy-2-{[3-(1-methylethyl)benzyl]amino}ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5- dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
6FTB
 
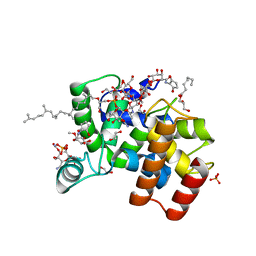 | | Staphylococcus aureus monofunctional glycosyltransferase in complex with moenomycin | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, 1,2-ETHANEDIOL, MOENOMYCIN, ... | | Authors: | Punekar, A.S, Dowson, C.J, Roper, D.I. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of the jaw subdomain of peptidoglycan glycosyltransferases for lipid II polymerization.
Cell Surf, 2, 2018
|
|
6FU1
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a n-alkyl hydroxamate | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | A Novel Class of Schistosoma mansoni Histone Deacetylase 8 (HDAC8) Inhibitors Identified by Structure-Based Virtual Screening and In Vitro Testing.
Molecules, 23, 2018
|
|
6GBO
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Ebola virus | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6FPL
 
 | | TETR(D) E147A MUTANT IN COMPLEX WITH TETRACYCLINE AND MAGNESIUM | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TETRACYCLINE, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2018-02-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
6FUN
 
 | |
6FS8
 
 | |
