6F2P
 
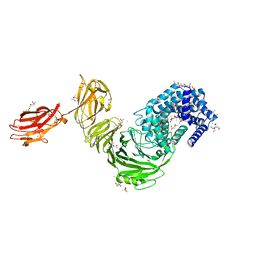 | | Structure of Paenibacillus xanthan lyase to 2.6 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lo Leggio, L, Kadziola, A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Dynamics of a Promiscuous Xanthan Lyase from Paenibacillus nanensis and the Design of Variants with Increased Stability and Activity.
Cell Chem Biol, 26, 2019
|
|
6EYM
 
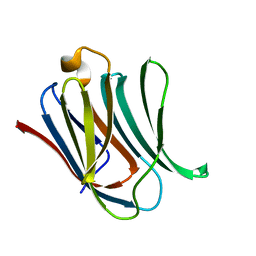 | | Neutron crystal structure of perdeuterated galectin-3C in complex with lactose | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Manzoni, F, Coates, L, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
6F6Y
 
 | |
6F28
 
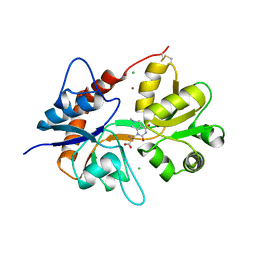 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2'-Amino-2'-carboxyethyl]-6-methyl-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.4A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
7Z1U
 
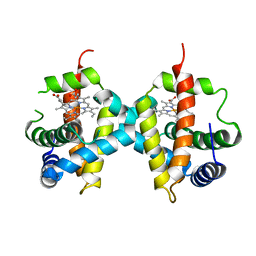 | | Biochemical implications of the substitution of a unique cysteine residue in sugar beet phytoglobin BvPgb 1.2 | | Descriptor: | Non-symbiotic hemoglobin class 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nyblom, M, Christensen, S, Leiva Eriksson, N, Bulow, L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
5DN8
 
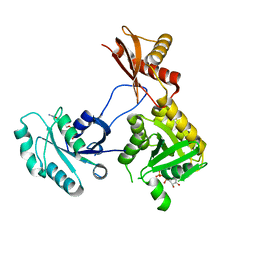 | | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP. | | Descriptor: | GTPase Der, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Minasov, G, Shuvalova, L, Han, A, Kim, H.-Y, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP.
To Be Published
|
|
5HLO
 
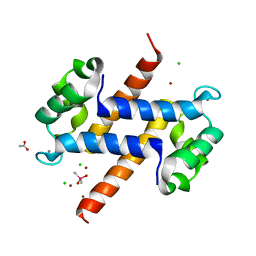 | | Crystal structure of calcium and zinc-bound human S100A8 in space group C2221 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
7Z2R
 
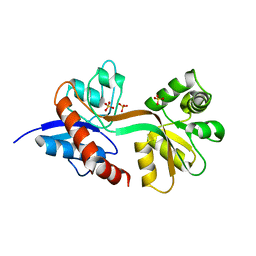 | | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics | | Descriptor: | Glutamate receptor ionotropic, delta-1, SULFATE ION | | Authors: | Masternak, M, Laulumaa, S, Kastrup, J.S. | | Deposit date: | 2022-02-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Differences between the GluD1 and GluD2 receptors revealed by GluD1 X-ray crystallography, binding studies and molecular dynamics.
Febs J., 290, 2023
|
|
5HLV
 
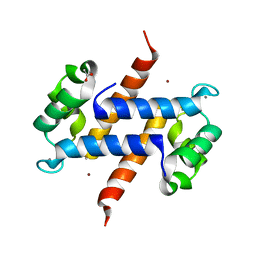 | | Crystal structure of calcium and zinc-bound human S100A8 in space group P212121 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
5E31
 
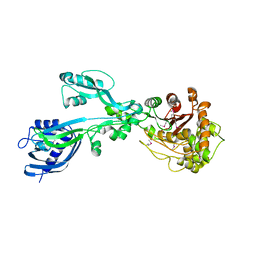 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5DVY
 
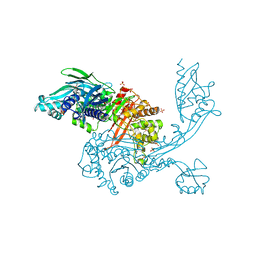 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
7Z3Y
 
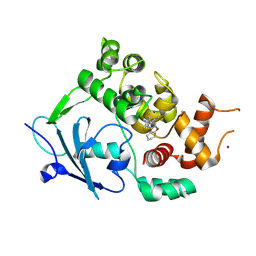 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)-2,6-bis(fluoranyl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Scaletti, E.R, Stenmark, P. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
5EQC
 
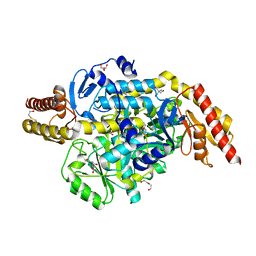 | | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site
To Be Published
|
|
5EYU
 
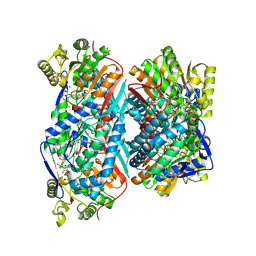 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5F2H
 
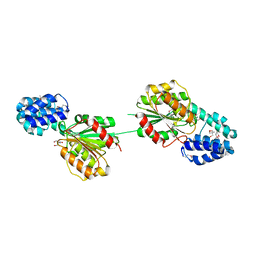 | | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Uncharacterized protein | | Authors: | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987
To Be Published
|
|
5SU5
 
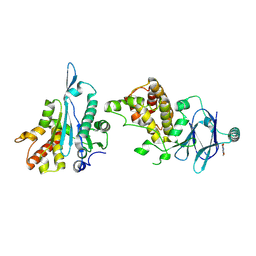 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03E05 from the F2X-Universal Library | | Descriptor: | 5-bromo-1-methylpyrimidine-2,4(1H,3H)-dione, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7ZOS
 
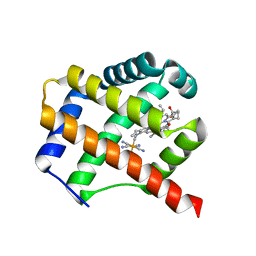 | | Class 1 Phytoglobin from Sugar beet (BvPgb1.2) | | Descriptor: | CYANIDE ION, HEXACYANOFERRATE(3-), Non-symbiotic hemoglobin class 1, ... | | Authors: | Nyblom, M, Christensen, S, Eriksson, N, Bulow, L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
7ZF0
 
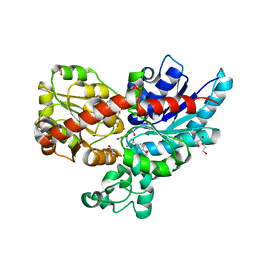 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP and p-hydroxymandelonitrile | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, ... | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
5IDI
 
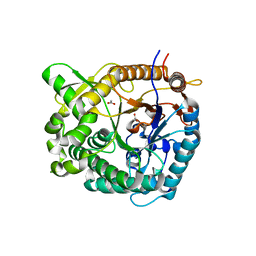 | | Structure of beta glucosidase 1A from Thermotoga neapolitana, mutant E349A | | Descriptor: | 1,4-beta-D-glucan glucohydrolase, ACETATE ION | | Authors: | Kulkarni, T, Nordberg Karlsson, E, Logan, D.T. | | Deposit date: | 2016-02-24 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-glucosidase 1A from Thermotoga neapolitana and comparison of active site mutants for hydrolysis of flavonoid glucosides.
Proteins, 85, 2017
|
|
7ZER
 
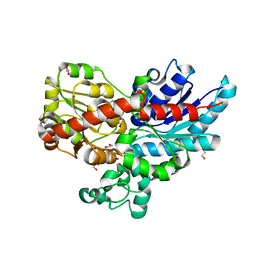 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZA4
 
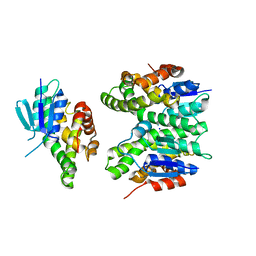 | | GSTF sh155 mutant | | Descriptor: | Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Directed Evolution of Phi Class Glutathione Transferases Involved in Multiple-Herbicide Resistance of Grass Weeds and Crops.
Int J Mol Sci, 23, 2022
|
|
5STN
 
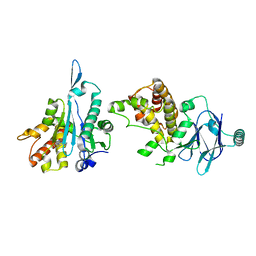 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03A11 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-methyl-1-{(1S,2R)-2-[(1,2,4-triazolidin-1-yl)methyl]cyclohexyl}methanamine, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
1W7H
 
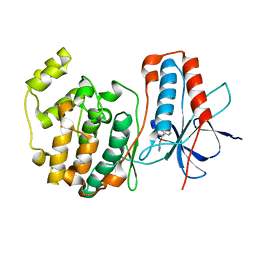 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Jhoti, H, Gill, A, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-02 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
5STT
 
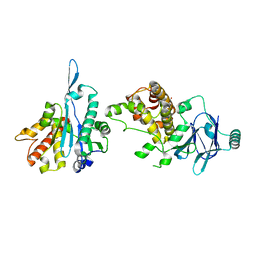 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03C03 from the F2X-Universal Library | | Descriptor: | 2-(2-methoxyethoxy)benzoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
5ST1
 
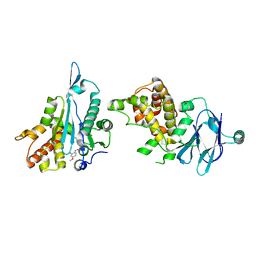 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P02C03 from the F2X-Universal Library | | Descriptor: | 5-methoxy-2-nitrosophenol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
