1O2D
 
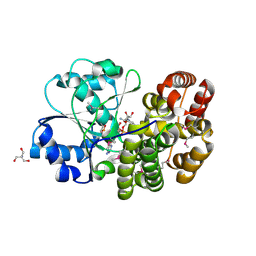 | |
6I8M
 
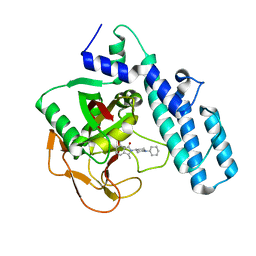 | |
2X2I
 
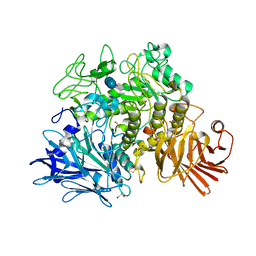 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
1HQS
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
3BHI
 
 | |
6JNK
 
 | |
5B3S
 
 | | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Shinzawa-Ito, K, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2016-03-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K)
To Be Published
|
|
7JHL
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP-N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
5UJB
 
 | | Structure of a Mcl-1 Inhibitor Binding to Site 3 of Human Serum Albumin | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, PHOSPHATE ION, Serum albumin | | Authors: | Zhao, B. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Myeloid cell leukemia-1 (Mcl-1) inhibitor bound to drug site 3 of Human Serum Albumin.
Bioorg. Med. Chem., 25, 2017
|
|
3QM3
 
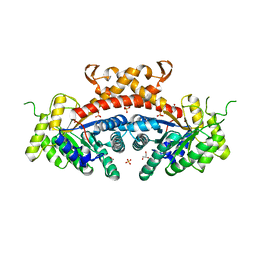 | | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, Fructose-bisphosphate aldolase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Fructose-bisphosphate Aldolase (Fba) from Campylobacter jejuni
TO BE PUBLISHED
|
|
1O9J
 
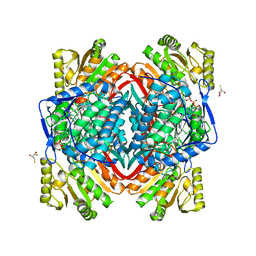 | | The X-ray crystal structure of eta-crystallin | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ALDEHYDE DEHYDROGENASE, ... | | Authors: | Purkiss, A.G, Van Montfort, R, Wistow, G, Slingsby, C. | | Deposit date: | 2002-12-15 | | Release date: | 2003-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Eta-Crystallin: Adaptation of a Class 1 Aldehyde Dehydrogenase for a New Role in the Eye Lens
Biochemistry, 42, 2003
|
|
2BHQ
 
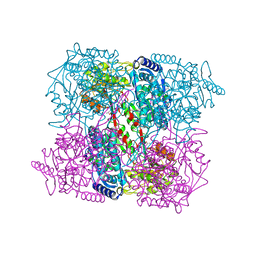 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound product glutamate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-01-16 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
7JHM
 
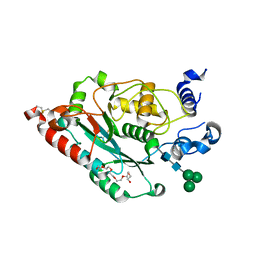 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
6AEL
 
 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
1JKF
 
 | | Holo 1L-myo-inositol-1-phosphate Synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, myo-inositol-1-phosphate synthase | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
1P1R
 
 | | Horse liver alcohol dehydrogenase complexed with NADH and R-N-1-methylhexylformamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R)-N-(1-METHYL-HEXYL)-FORMAMIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Venkataramaiah, T.H, Plapp, B.V. | | Deposit date: | 2003-04-13 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Formamides mimic aldehydes and inhibit liver alcohol dehydrogenases and ethanol metabolism
J.Biol.Chem., 278, 2003
|
|
6JNJ
 
 | |
4K57
 
 | |
1I1X
 
 | |
1QAS
 
 | |
4HFR
 
 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with an orally bioavailable acidic inhibitor AZD4017. | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {(3S)-1-[5-(cyclohexylcarbamoyl)-6-(propylsulfanyl)pyridin-2-yl]piperidin-3-yl}acetic acid | | Authors: | Ogg, D.J, Gerhardt, S, Hargreaves, D. | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Bioavailable Acidic 11 -Hydroxysteroid Dehydrogenase Type 1 (11 -HSD1) Inhibitor: Discovery of 2-[(3S)-1-[5-(Cyclohexylcarbamoyl)-6-propylsulfanylpyridin-2-yl]-3-piperidyl]acetic Acid (AZD4017)
J.Med.Chem., 55, 2012
|
|
4AMX
 
 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-glucosyl- fluoride | | Descriptor: | 5-fluoro-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
1RE9
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450-CAM WITH A FLUORESCENT PROBE D-8-AD (ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-06 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires.
J.Mol.Biol., 344, 2004
|
|
1JFX
 
 | | Crystal structure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolution | | Descriptor: | 1,4-beta-N-Acetylmuramidase M1, CHLORIDE ION | | Authors: | Rau, A, Hogg, T, Marquardt, R, Hilgenfeld, R. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new lysozyme fold. Crystal structure of the muramidase from Streptomyces coelicolor at 1.65 A resolution.
J.Biol.Chem., 276, 2001
|
|
2FUR
 
 | |
