2MSC
 
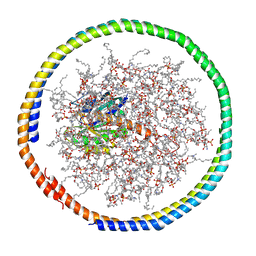 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2JZD
 
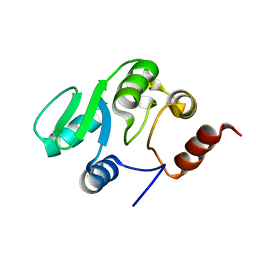 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2JZE
 
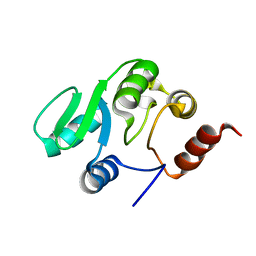 | | NMR structure of the domain 527-651 of the SARS-CoV nonstructural protein nsp3, single conformer closest to the mean coordinates of an ensemble of twenty energy minimized conformers | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2POJ
 
 | |
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2MMA
 
 | | NMR-based docking model of GrxS14-BolA2 apo-heterodimer from Arabidopsis thaliana | | Descriptor: | BolA2, Monothiol glutaredoxin-S14, chloroplastic | | Authors: | Roret, T, Tsan, P, Couturier, J, Rouhier, N, Didierjean, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Spectroscopic Insights into BolA-Glutaredoxin Complexes.
J.Biol.Chem., 289, 2014
|
|
1XPN
 
 | | NMR structure of P. aeruginosa protein PA1324: Northeast Structural Genomics Consortium target PaP1 | | Descriptor: | hypothetical protein PA1324 | | Authors: | Cort, J.R, Ni, S, Lockert, E.E, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Structure and function of Pseudomonas aeruginosa protein PA1324 (21-170).
Protein Sci., 18, 2009
|
|
2MRY
 
 | |
2L4W
 
 | | NMR structure of the Xanthomonas VirB7 | | Descriptor: | Uncharacterized protein | | Authors: | Souza, D.P, Farah, C.S, Salinas, R.K. | | Deposit date: | 2010-10-15 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins.
Plos Pathog., 7, 2011
|
|
2LPI
 
 | | NMR structure of a monomeric mutant (A72R) of major ampullate spidroin 1 N-terminal domain | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S.D, Johansson, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain.
J.Mol.Biol., 422, 2012
|
|
2LPJ
 
 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 7.2 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S.D, Johansson, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain.
J.Mol.Biol., 422, 2012
|
|
2BTB
 
 | |
2BTA
 
 | |
1XRZ
 
 | | NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue | | Descriptor: | ZINC ION, Zinc finger Y-chromosomal protein | | Authors: | Lachenmann, M.J, Ladbury, J.E, Qian, X, Huang, K, Singh, R, Weiss, M.A. | | Deposit date: | 2004-10-17 | | Release date: | 2004-11-30 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solvation and the hidden thermodynamics of a zinc finger probed
by nonstandard repair of a protein crevice
Protein Sci., 13, 2004
|
|
2MAW
 
 | |
2L9F
 
 | | NMR solution structure of meACP | | Descriptor: | CalE8 | | Authors: | Lim, J, Yang, D, Liang, Z.X, Kong, R, Murugan, E, Ho, C.L. | | Deposit date: | 2011-02-08 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8
Plos One, 6, 2011
|
|
2NA0
 
 | |
2LQR
 
 | | NMR structure of Ig3 domain of palladin | | Descriptor: | Palladin | | Authors: | Beck, M.R, Dixon IV, R.D.S, Otey, C.A, Campbell, S.L, Murphy, G.S. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of Palladin's Actin Binding Domain.
J.Mol.Biol., 425, 2013
|
|
1X5V
 
 | | NMR Structure of PcFK1 | | Descriptor: | PcFK1 | | Authors: | Pimentel, C, Choi, S.J, Chagot, B, Guette, C, Camadro, J.M, Darbon, H. | | Deposit date: | 2005-05-17 | | Release date: | 2006-04-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PcFK1, a spider peptide active against Plasmodium falciparum
Protein Sci., 15, 2006
|
|
2MLX
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA220-310 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2LVS
 
 | |
2M5I
 
 | |
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
2NZ3
 
 | |
2NY9
 
 | |
