7G3S
 
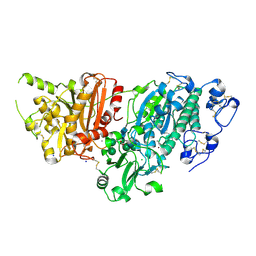 | | Crystal Structure of rat Autotaxin in complex with 2-[(5-chloro-2,3-dihydro-1H-inden-2-yl)amino]-6-(oxetan-3-yl)-7H-pyrrolo[3,4-d]pyrimidin-5-one, i.e. SMILES O=C1N(C2COC2)Cc2nc(N[C@H]3Cc4c(C3)ccc(c4)Cl)ncc12 with IC50=0.150181 microM | | Descriptor: | 2-{[(2S)-5-chloro-2,3-dihydro-1H-inden-2-yl]amino}-6-(oxetan-3-yl)-6,7-dihydro-5H-pyrrolo[3,4-d]pyrimidin-5-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Pinard, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G3D
 
 | | Crystal Structure of rat Autotaxin in complex with 4-tert-butyl-2-chloro-5-(1H-pyrazolo[3,4-b]pyridin-3-ylmethoxy)benzonitrile, i.e. SMILES CC(C)(C)c1cc(Cl)c(C#N)cc1OCC1=NNc2ncccc12 with IC50=0.0201187 microM | | Descriptor: | 4-tert-butyl-2-chloro-5-[(1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]benzonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2VR2
 
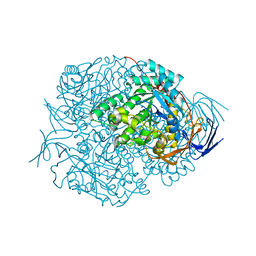 | | Human Dihydropyrimidinase | | Descriptor: | CHLORIDE ION, DIHYDROPYRIMIDINASE, ZINC ION | | Authors: | Welin, M, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Human Dihydropyrimidinase
To be Published
|
|
7G3K
 
 | | Crystal Structure of rat Autotaxin in complex with 5-tert-butyl-2-chloro-4-[(7-methyl-8-oxo-5,6-dihydroimidazo[1,5-a]pyrazin-3-yl)methoxy]benzonitrile, i.e. SMILES c1c(c(cc(c1C(C)(C)C)OCC1=NC=C2N1CCN(C2=O)C)Cl)C#N with IC50=0.416172 microM | | Descriptor: | 5-tert-butyl-2-chloro-4-{[(4S)-7-methyl-8-oxo-5,6,7,8-tetrahydroimidazo[1,5-a]pyrazin-3-yl]methoxy}benzonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Green, L, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G3X
 
 | | Crystal Structure of rat Autotaxin in complex with (2S)-1-[2-(2,1,3-benzothiadiazol-4-yloxy)acetyl]azetidine-2-carboxamide | | Descriptor: | (2R)-1-{[(2,1,3-benzothiadiazol-4-yl)oxy]acetyl}azetidine-2-carboxamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Cai, J, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G3W
 
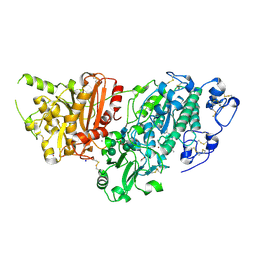 | | Crystal Structure of rat Autotaxin in complex with N-[(2R)-5-chloro-2,3-dihydro-1H-inden-2-yl]-5-(3-methyl-1,2,4-oxadiazol-5-yl)pyrimidin-2-amine, i.e. SMILES c1c(cnc(n1)N[C@H]1Cc2cc(Cl)ccc2C1)C1=NC(=NO1)C with IC50=0.186827 microM | | Descriptor: | ACETATE ION, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Pinard, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G45
 
 | | Crystal Structure of rat Autotaxin in complex with 3-(4-chloro-5-methyl-2-propan-2-ylphenyl)-4-methyl-1H-1,2,4-triazol-5-one, i.e. SMILES c1c(c(cc(c1C(C)C)C1=NNC(=O)N1C)C)Cl with IC50=0.843581 microM | | Descriptor: | (5M)-5-[4-chloro-5-methyl-2-(propan-2-yl)phenyl]-4-methyl-2,4-dihydro-3H-1,2,4-triazol-3-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G48
 
 | | Crystal Structure of rat Autotaxin in complex with [(3aS,6aS)-5-(4-methoxynaphthalene-2-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-(1H-benzotriazol-5-yl)methanone, i.e. SMILES N1(C[C@@H]2[C@@H](C1)CN(C2)C(=O)c1cc(c2c(c1)cccc2)OC)C(=O)c1ccc2c(c1)N=NN2 with IC50=0.11329 microM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
4O6V
 
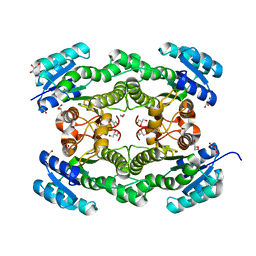 | |
7G4A
 
 | | Crystal Structure of rat Autotaxin in complex with (3-chloro-5-methylsulfonylphenyl)methyl 2-(1H-benzotriazole-5-carbonyl)-2,7-diazaspiro[3.5]nonane-7-carboxylate, i.e. SMILES c1c(cc(cc1S(=O)(=O)C)COC(=O)N1CCC2(CC1)CN(C2)C(=O)c1ccc2c(c1)N=NN2)Cl with IC50=0.332347 microM | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2G17
 
 | |
7G4F
 
 | | Crystal Structure of rat Autotaxin in complex with (E)-1-[(3aR,6aR)-5-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-3-[4-(trifluoromethoxy)phenyl]prop-2-en-1-one, i.e. SMILES C1N(C[C@H]2[C@H]1CN(C2)C(=O)c1ccc2c(c1)N=NN2)C(=O)/C=C/c1ccc(cc1)OC(F)(F)F with IC50=0.00118848 microM | | Descriptor: | (2E)-1-[(3aR,6aS)-5-(1H-benzotriazole-5-carbonyl)hexahydropyrrolo[3,4-c]pyrrol-2(1H)-yl]-3-[4-(trifluoromethoxy)phenyl]prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
6IHD
 
 | | Crystal structure of Malate dehydrogenase from Metallosphaera sedula | | Descriptor: | 2-ETHOXYETHANOL, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, D, Kim, K.J. | | Deposit date: | 2018-09-29 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and biochemical characterization of malate dehydrogenase from Metallosphaera sedula
Biochem. Biophys. Res. Commun., 509, 2019
|
|
7G4J
 
 | | Crystal Structure of rat Autotaxin in complex with 4-[rac-(E)-3-oxo-3-[rac-(3aR,8aS)-2-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,5,7,8,8a-octahydropyrrolo[3,4-d]azepin-6-yl]prop-1-enyl]benzonitrile, i.e. SMILES C1C[C@@H]2[C@H](CCN1C(=O)/C=C/c1ccc(cc1)C#N)CN(C2)C(=O)c1ccc2c(c1)N=NN2 with IC50=0.193037 microM | | Descriptor: | 4-{(1E)-3-[(3aR,8aS)-2-(1H-benzotriazole-5-carbonyl)octahydropyrrolo[3,4-d]azepin-6(1H)-yl]-3-oxoprop-1-en-1-yl}benzonitrile, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G4O
 
 | | Crystal Structure of rat Autotaxin in complex with [3-(2-hydroxy-4,6-dimethylphenyl)-3-methylbutyl] 2,2-dimethylpropanoate, i.e. SMILES c1c(cc(c(c1O)C(CCOC(=O)C(C)(C)C)(C)C)C)C with IC50=1.13176 microM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(2-hydroxy-4,6-dimethylphenyl)-3-methylbutyl 2,2-dimethylpropanoate, ACETATE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mizuguchi, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2OGR
 
 | |
5ZJE
 
 | | LDHA-mla | | Descriptor: | L-lactate dehydrogenase A chain, MALONATE ION | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.929 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
4C7L
 
 | | Crystal structure of Mouse Hepatitis virus strain S Hemagglutinin- esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zeng, Q.H, Huizinga, E.G. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Murine Coronavirus Hemagglutinin-Esterase Receptor-Binding Site: A Major Shift in Ligand Specificity Through Modest Changes in Architecture.
Plos Pathog., 8, 2012
|
|
6IMO
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[(1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
4LA4
 
 | |
3I6Y
 
 | | Structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
2GC2
 
 | |
2W67
 
 | | BtGH84 in complex with FMA34 | | Descriptor: | CALCIUM ION, GLYCEROL, N-[(3S,4R,5R,6R)-4,5,6-trihydroxyazepan-3-yl]acetamide, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2008-12-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Basis for Inhibition of Gh84 Glycoside Hydrolases by Substituted Azepanes: Conformational Flexibility Enables Probing of Substrate Distortion.
J.Am.Chem.Soc., 131, 2009
|
|
4OCL
 
 | |
