2CZ3
 
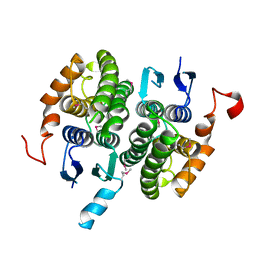 | | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-2 crystal) | | Descriptor: | Maleylacetoacetate isomerase | | Authors: | Mizohata, E, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-2 crystal)
To be Published
|
|
2CZ4
 
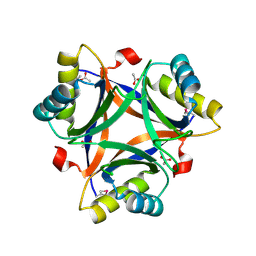 | | Crystal structure of a putative PII-like signaling protein (TTHA0516) from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, hypothetical protein TTHA0516 | | Authors: | Arai, R, Fusatomi, E, Kukimoto-Niino, M, Kawaguchi, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of a putative PII-like signaling protein (TTHA0516) from Thermus thermophilus HB8
To be Published
|
|
2CZ5
 
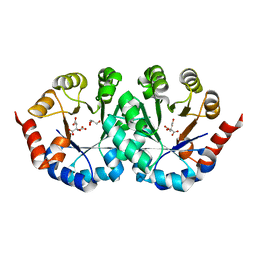 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3
To be Published
|
|
2CZ6
 
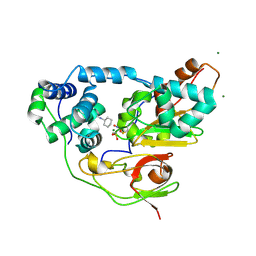 | | Complex of Inactive Fe-type NHase with Cyclohexyl isocyanide | | Descriptor: | CYCLOHEXYL ISOCYANIDE, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Nojiri, M, Kawano, Y, Hashimoto, K, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | x-ray snap shots of inhibitor binding process in photo-reactive nitrile hydratase
To be Published
|
|
2CZ7
 
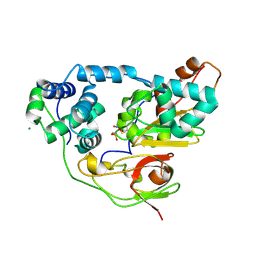 | | Fe-type NHase photo-activated for 75min at 105K | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Nojiri, M, Kawano, Y, Hashimoto, K, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active Center Structures of Photo-reactive Nitrile Hydratase during its Photo-activation Process
To be Published
|
|
2CZ8
 
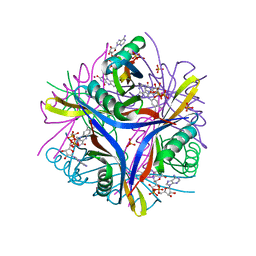 | | Crystal Structure of tt0972 protein from Thermus thermophilus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kumei, M, Inagaki, E, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of tt0972 protein from Thermus thermophilus
To be Published
|
|
2CZ9
 
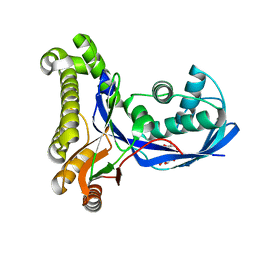 | | Crystal Structure of galactokinase from Pyrococcus horikoshi | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable galactokinase | | Authors: | Inagaki, E, Sakamoto, K, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of galactokinase from Pyrococcus horikoshi
To be Published
|
|
2CZC
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Pyrococcus horikoshii OT3 | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Ito, K, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Pyrococcus horikoshii OT3
To be Published
|
|
2CZD
 
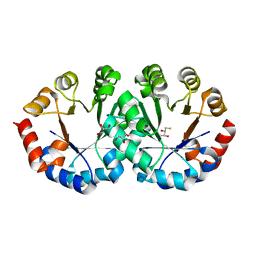 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 at 1.6 A resolution
To be Published
|
|
2CZE
 
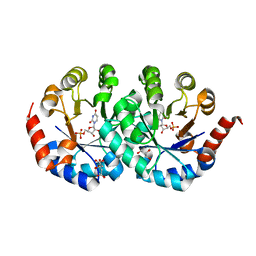 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Arai, R, Ito, K, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 complexed with UMP
To be Published
|
|
2CZF
 
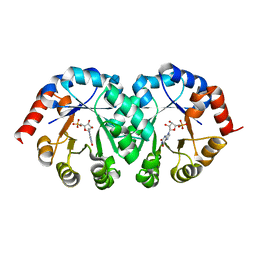 | |
2CZG
 
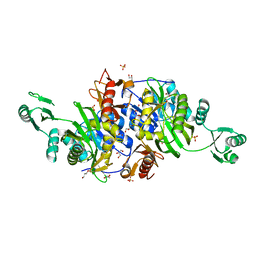 | | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyl transferase | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3
To be Published
|
|
2CZH
 
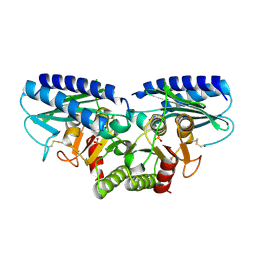 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) with phosphate ion (orthorhombic form) | | Descriptor: | Inositol monophosphatase 2, PHOSPHATE ION | | Authors: | Ito, K, Arai, R, Kamo-Uchikubo, T, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2CZI
 
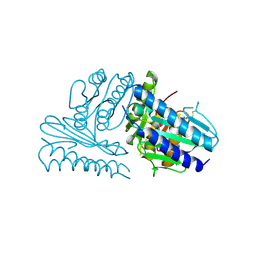 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) with calcium and phosphate ions | | Descriptor: | CALCIUM ION, Inositol monophosphatase 2, PHOSPHATE ION | | Authors: | Arai, R, Ito, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2CZJ
 
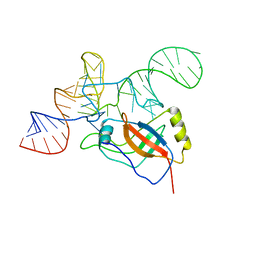 | | Crystal structure of the tRNA domain of tmRNA from Thermus thermophilus HB8 | | Descriptor: | SsrA-binding protein, tmRNA (63-MER) | | Authors: | Bessho, Y, Shibata, R, Sekine, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2CZK
 
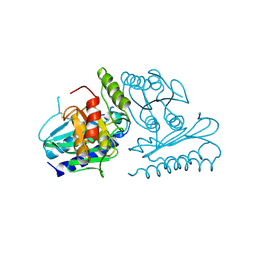 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (trigonal form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Hanawa-Suetsugu, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
2CZL
 
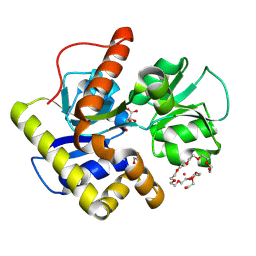 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 (Cys11 modified with beta-mercaptoethanol) | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Kamo-Uchikubo, T, Nishimoto, M, Toyama, M, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
2CZN
 
 | | Solution structure of the chitin-binding domain of hyperthermophilic chitinase from pyrococcus furiosus | | Descriptor: | chitinase | | Authors: | Uegaki, T, Ikegami, T, Nakamura, T, Hagihara, Y, Mine, S, Inoue, T, Matsumura, H, Ataka, M, Ishikawa, K. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Mol.Biol., 381, 2008
|
|
2CZO
 
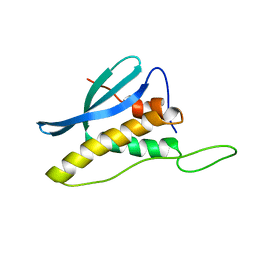 | | Solution Structure of the PX Domain of Bem1p | | Descriptor: | Bud emergence protein 1 | | Authors: | Maeda, A, Ogura, K, Horiuchi, M, Kumeta, H, Fujioka, Y, Inagaki, F. | | Deposit date: | 2005-07-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PX domain of Bem1p
To be Published
|
|
2CZP
 
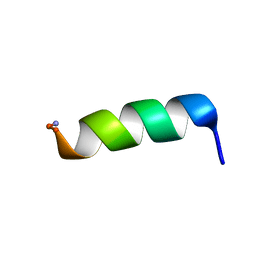 | | Structural analysis of membrane-bound mastoparan-X by solid-state NMR | | Descriptor: | Mastoparan X | | Authors: | Todokoro, Y, Fujiwara, T, Yumen, I, Fukushima, K, Kang, S.-W, Park, J.-S, Kohno, T, Wakamatsu, K, Akutsu, H. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-13 | | Method: | SOLID-STATE NMR | | Cite: | Structure of tightly membrane-bound mastoparan-x, a g-protein-activating Peptide, determined by solid-state NMR.
Biophys.J., 91, 2006
|
|
2CZQ
 
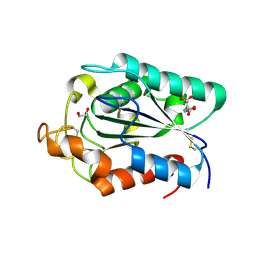 | | A novel cutinase-like protein from Cryptococcus sp. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, cutinase-like protein | | Authors: | Masaki, K, Kamini, N.R, Ikeda, H, Iefuji, H, Kondo, H, Suzuki, M, Tsuda, S. | | Deposit date: | 2005-07-14 | | Release date: | 2006-07-14 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2
Proteins, 77, 2009
|
|
2CZR
 
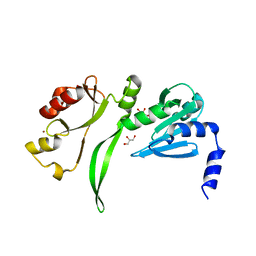 | | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA | | Descriptor: | GLYCEROL, TBP-interacting protein, ZINC ION | | Authors: | Yamamoto, T, Matsuda, T, Inoue, T, Matsumura, H, Morikawa, M, Kanaya, S, Kai, Y. | | Deposit date: | 2005-07-15 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA
Protein Sci., 15, 2006
|
|
2CZS
 
 | |
2CZT
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
2CZU
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
