3PMI
 
 | | PWWP Domain of Human Mutated Melanoma-Associated Antigen 1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PWWP domain-containing protein MUM1, SULFATE ION, ... | | Authors: | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
6TTZ
 
 | | Structure of the ClpP:ADEP4-complex from Staphylococcus aureus (open state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, N-[(2S)-3-(3,5-difluorophenyl)-1-[[(3S,9S,13S,15R,19S,22S)-15,19-dimethyl-2,8,12,18,21-pentaoxo-11-oxa-1,7,17,20-tetrazatetracyclo[20.4.0.03,7.013,17]hexacosan-9-yl]amino]-1-oxopropan-2-yl]heptanamide | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
6TTY
 
 | | Structure of ClpP from Staphylococcus aureus (apo, closed state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
2LP0
 
 | |
6J93
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase form apo at 0.95 A resolution.
To Be Published
|
|
6JKX
 
 | | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution. | | Descriptor: | CHLORIDE ION, METHANOL, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution.
To Be Published
|
|
6JJ1
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-24 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues
To Be Published
|
|
6JGU
 
 | |
6JJQ
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Bairagya, H.R, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution.
To Be Published
|
|
3PFS
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 3 | | Descriptor: | Bromodomain and PHD finger-containing protein 3, SULFATE ION, ZINC ION | | Authors: | Lam, R, Zeng, H, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
2FOT
 
 | | Crystal structure of the complex between calmodulin and alphaII-spectrin | | Descriptor: | CALCIUM ION, Calmodulin, alpha-II spectrin Spectrin | | Authors: | Simonovic, M, Zhang, Z, Cianci, C.D, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2006-01-13 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the calmodulin alphaII-spectrin complex provides insight into the regulation of cell plasticity.
J.Biol.Chem., 281, 2006
|
|
6PH4
 
 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH3
 
 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PPS
 
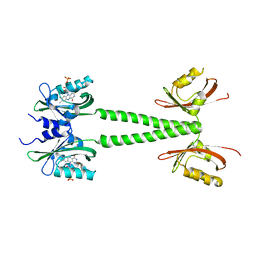 | | A blue light illuminated LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Fernandez, I, Shin, H, Gunawardana, S, Otero, L.H, Cerutti, M.L, Yang, X, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH2
 
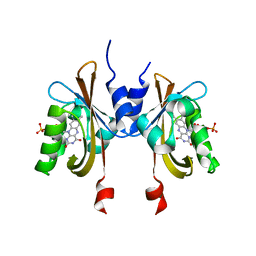 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
4BIW
 
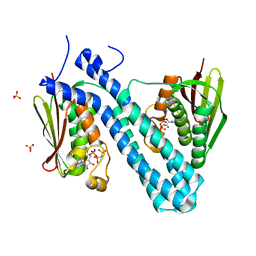 | | Crystal structure of CpxAHDC (hexagonal form) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SENSOR PROTEIN CPXA, ... | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIY
 
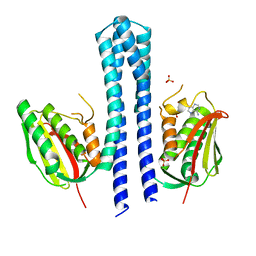 | | Crystal structure of CpxAHDC (monoclinic form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIU
 
 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIX
 
 | | Crystal structure of CpxAHDC (monoclinic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SENSOR PROTEIN CPXA, ... | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIV
 
 | | Crystal structure of CpxAHDC (trigonal form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SENSOR PROTEIN CPXA | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
3TYY
 
 | | Crystal Structure of Human Lamin-B1 Coil 2 Segment | | Descriptor: | Lamin-B1 | | Authors: | Lam, R, Xu, C, Bian, C.B, Mackenzie, F, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structures of the coil 2B fragment and the globular tail domain of human lamin B1.
Febs Lett., 586, 2012
|
|
3V30
 
 | | Crystal Structure of the Peptide Bound Complex of the Ankyrin Repeat Domains of Human RFXANK | | Descriptor: | DNA-binding protein RFX5, DNA-binding protein RFXANK | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
5AAR
 
 | |
2VX9
 
 | | H. salinarum dodecin E45A mutant | | Descriptor: | CHLORIDE ION, DODECIN, RIBOFLAVIN, ... | | Authors: | Grininger, M, Staudt, H, Johansson, P, Wachtveitl, J, Oesterhelt, D. | | Deposit date: | 2008-07-01 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dodecin is the Key Player in Flavin Homeostasis of Archaea.
J.Biol.Chem., 284, 2009
|
|
2VXA
 
 | | H. halophila dodecin in complex with riboflavin | | Descriptor: | CHLORIDE ION, DODECIN, RIBOFLAVIN | | Authors: | Grininger, M, Staudt, H, Johansson, P, Wachtveitl, J, Oesterhelt, D. | | Deposit date: | 2008-07-01 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dodecin is the Key Player in Flavin Homeostasis of Archaea.
J.Biol.Chem., 284, 2009
|
|
