2BI2
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure C) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI3
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure D) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI4
 
 | | Lactaldehyde:1,2-propanediol oxidoreductase of Escherichia coli | | Descriptor: | CHLORIDE ION, FE (III) ION, LACTALDEHYDE REDUCTASE, ... | | Authors: | Montella, C, Bellsolell, L, Badia, J, Baldoma, L, Perez, R, Coll, M, Aguilar, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of an Iron-Dependent Group III Dehydrogenase that Interconverts L-Lactaldehyde and L-1,2-Propanediol in Escherichia Coli
J.Bacteriol., 187, 2005
|
|
2BI5
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure E) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI6
 
 | | NMR STUDY OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
2BI7
 
 | | udp-galactopyranose mutase from Klebsiella pneumoniae oxidised FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Srikannathasan, V, Naismith, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mycobacteria Tuberculosis and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the Oxidised State and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the (Active) Reduced State.
J.Mol.Biol., 348, 2005
|
|
2BI8
 
 | |
2BI9
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure F) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIA
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure G) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIB
 
 | | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PHOSPHOCHOLINE, ... | | Authors: | Hermoso, J.A, Lagartera, L, Gonzalez, A, Garcia, P, Martinez-Ripoll, M, Garcia, J.L, Menendez, M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights Into Pneumococcal Pathogenesis from Crystal Structure of the Modular Teichoic Acid Phosphorylcholine Esterase Pce
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BIC
 
 | | The solution structure of the recombinant elicitor protein PcF from the oomycete pathogen P. cactorum | | Descriptor: | PHYTOTOXIC PROTEIN PCF | | Authors: | Nicastro, G, Orsomando, G, Desario, F, Ferrari, E, Manconi, L, Spisni, A, Ruggieri, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-06-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Phytotoxic Protein Pcf: The First Characterized Member of the Phytophthora Pcf Toxin Family.
Protein Sci., 18, 2009
|
|
2BID
 
 | | HUMAN PRO-APOPTOTIC PROTEIN BID | | Descriptor: | PROTEIN (BID) | | Authors: | Chou, J.J, Li, H, Salvesen, G.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-01-27 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BID, an intracellular amplifier of apoptotic signaling.
Cell(Cambridge,Mass.), 96, 1999
|
|
2BIE
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure H) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIF
 
 | | 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE H256A MUTANT WITH F6P IN PHOSPHATASE ACTIVE SITE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Yuen, M.H, Hasemann, C.A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities.
J.Biol.Chem., 274, 1999
|
|
2BIG
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure I) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIH
 
 | | crystal structure of the Molybdenum-containing nitrate reducing fragment of Pichia angusta assimilatory nitrate reductase | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(IV), NITRATE REDUCTASE [NADPH] | | Authors: | Fischer, K, Barbier, G, Hecht, H.-J, Mendel, R.R, Campbell, W.H, Schwarz, G. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Eukaryotic Nitrate Reduction: Crystal Structures of the Nitrate Reductase Active Site
Plant Cell, 17, 2005
|
|
2BII
 
 | | crystal structure of nitrate-reducing fragment of assimilatory nitrate reductase from Pichia angusta | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(IV), GLYCEROL, NITRATE REDUCTASE [NADPH], ... | | Authors: | Fischer, K, Barbier, G, Hecht, H.-J, Mendel, R.R, Campbell, W.H, Schwarz, G. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Eukaryotic Nitrate Reduction: Crystal Structures of the Nitrate Reductase Active Site
Plant Cell, 17, 2005
|
|
2BIJ
 
 | | Crystal structure of the human protein tyrosine phosphatase PTPN5 (STEP, striatum enriched enriched Phosphatase) | | Descriptor: | SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE, NON-RECEPTOR TYPE 5 | | Authors: | Barr, A.J, Debreczeni, J.E, Eswaran, J, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem. J., 395, 2006
|
|
2BIK
 
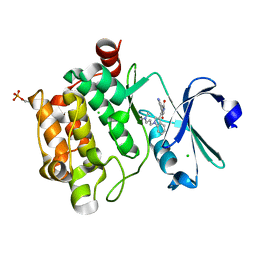 | | Human Pim1 phosphorylated on Ser261 | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, CHLORIDE ION, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K. | | Deposit date: | 2005-01-22 | | Release date: | 2005-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Pim1 Phosphorylated on Ser261
To be Published
|
|
2BIL
 
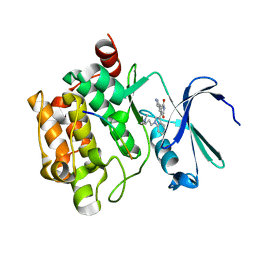 | | The human protein kinase Pim1 in complex with its consensus peptide Pimtide | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, CONSENSUS PIM1 PEPTIDE PIMTIDE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K. | | Deposit date: | 2005-01-22 | | Release date: | 2005-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Human Protein Kinase Pim1 in Complex with its Consensus Peptide Pimtide
To be Published
|
|
2BIM
 
 | |
2BIN
 
 | |
2BIO
 
 | |
2BIP
 
 | |
2BIQ
 
 | |
