7YOF
 
 | |
6J7U
 
 | |
7YOG
 
 | |
7YOI
 
 | |
7YOK
 
 | | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A | | Descriptor: | Cysteine synthase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A
To Be Published
|
|
7YOL
 
 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
7YOD
 
 | |
5WC8
 
 | |
7YOH
 
 | |
7YOM
 
 | | Crystal structure of tetra mutant (D67E,A68P,L98I,A301S) of O-acetylserine sulfhydrylase from Salmonella typhimurium in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.8 A | | Descriptor: | Cysteine synthase, peptide from serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of I88L single mutant of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.14 A
To Be Published
|
|
5WC6
 
 | |
7YVY
 
 | |
5WJD
 
 | | Crystal structure of Naa80 bound to acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, CG8481, isoform B, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2GG4
 
 | | CP4 EPSP synthase (unliganded) | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Schonbrunn, E, Funke, T. | | Deposit date: | 2006-03-23 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the herbicide resistance of Roundup Ready crops.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6M31
 
 | |
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
7ZG5
 
 | | The crystal structure of Salmonella TacAT3-DNA complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetyltransferase, BARIUM ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S. | | Deposit date: | 2022-04-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular stripping underpins derepression of a toxin-antitoxin system.
Nat.Struct.Mol.Biol., 2024
|
|
6LOE
 
 | | Cryo-EM structure of the dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii | | Descriptor: | Cytochrome c domain-containing protein, FE3-S4 CLUSTER, Fe-S-cluster-containing hydrogenase components 1-like protein, ... | | Authors: | Shi, Y, Xin, Y.Y, Wang, C, Blankenship, R.E, Sun, F, Xu, X.L. | | Deposit date: | 2020-01-05 | | Release date: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the air-oxidized and dithionite-reduced photosynthetic alternative complex III from Roseiflexus castenholzii .
Sci Adv, 6, 2020
|
|
6LQV
 
 | |
4PSW
 
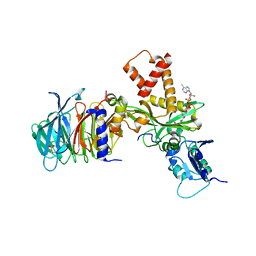 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
6LOI
 
 | |
2GL1
 
 | | NMR solution structure of Vigna radiata Defensin 2 (VrD2) | | Descriptor: | PDF1 | | Authors: | Lin, K.F, Lee, T.R, Tsai, P.H, Hsu, M.P, Chen, C.S, Lyu, P.C. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-based protein engineering for alpha-amylase inhibitory activity of plant defensin.
Proteins, 68, 2007
|
|
7ZP0
 
 | |
6LQQ
 
 | |
