1ECL
 
 | |
201D
 
 | |
1GNR
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
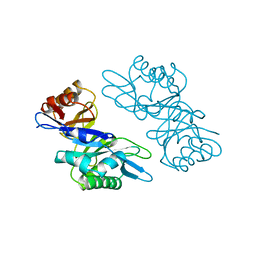
1EDT
 
 | |
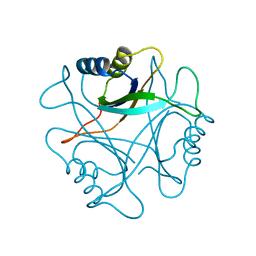
1PIL
 
 | |
1ENX
 
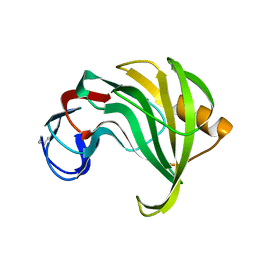 | |
1XYP
 
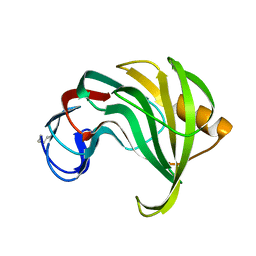 | |
1XYN
 
 | |
1XYO
 
 | |
3PTE
 
 | |
1STD
 
 | |
1BNC
 
 | |
1SMV
 
 | |
1MNP
 
 | | MANGANESE PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Sundaramoorthy, M, Poulos, T.L. | | Deposit date: | 1995-01-27 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preliminary crystallographic analysis of manganese peroxidase from Phanerochaete chrysosporium.
J.Mol.Biol., 238, 1994
|
|
1HLS
 
 | | NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) | | Descriptor: | INSULIN | | Authors: | Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1995-06-28 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an engineered biologically potent insulin monomer, B16 Tyr-->His, as determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1DIH
 
 | |
1DMA
 
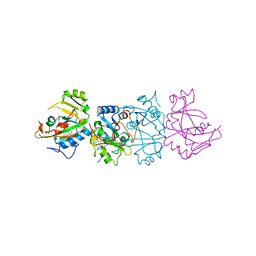 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH NICOTINAMIDE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, EXOTOXIN A, NICOTINAMIDE | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D. | | Deposit date: | 1995-04-28 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Pseudomonas aeruginosa exotoxin domain III with nicotinamide and AMP: conformational differences with the intact exotoxin.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1DCT
 
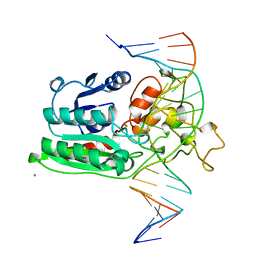 | | DNA (CYTOSINE-5) METHYLASE FROM HAEIII COVALENTLY BOUND TO DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*CP*AP*GP*CP*AP*GP*GP*(C49)P*CP*AP*CP*CP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*TP*GP*GP*(C5M)P*CP*TP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Reinisch, K.M, Chen, L, Verdine, G.L, Lipscomb, W.N. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing.
Cell(Cambridge,Mass.), 82, 1995
|
|
1MTI
 
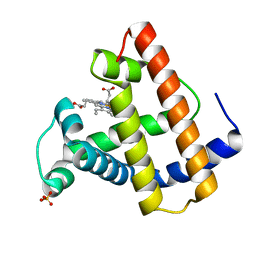 | |
1MTJ
 
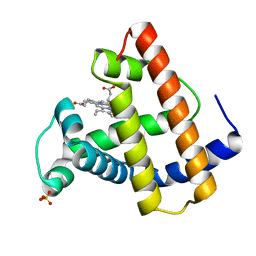 | |
1TTJ
 
 | |
1ORD
 
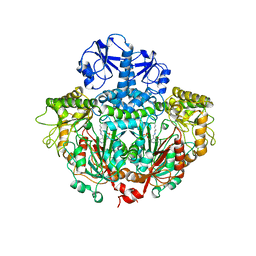 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
1IDM
 
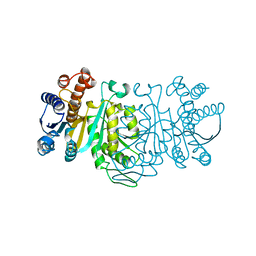 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1OVT
 
 | |
