1RCK
 
 | |
2IFO
 
 | |
1ENJ
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1RCL
 
 | |
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
2PEC
 
 | |
1XYP
 
 | |
1XYN
 
 | |
1XYO
 
 | |
1MDP
 
 | |
1MDQ
 
 | |
185D
 
 | |
184D
 
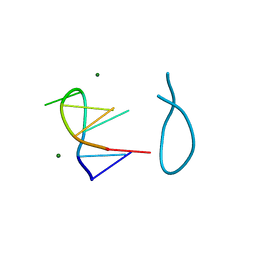 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
1EAP
 
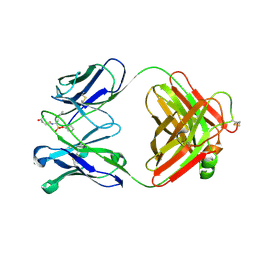 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
1HNG
 
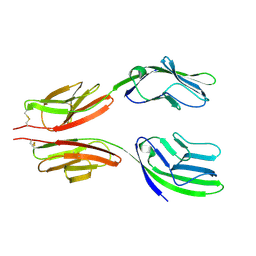 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
1MPF
 
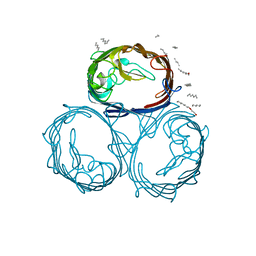 | |
1HNF
 
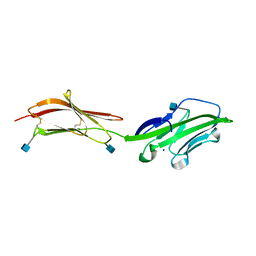 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
1RLR
 
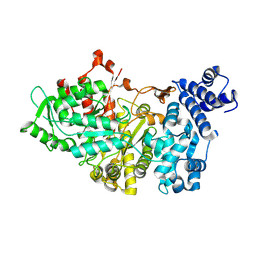 | |
1GBR
 
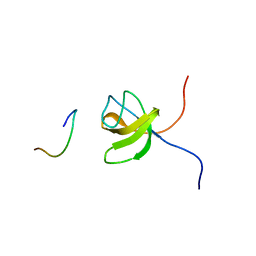 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | Authors: | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | Deposit date: | 1994-08-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
2PTL
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN-BINDING DOMAIN OF PROTEIN L. COMPARISON WITH THE IGG-BINDING DOMAINS OF PROTEIN G | | Descriptor: | PROTEIN L | | Authors: | Wikstroem, M, Drakenberg, T, Forsen, S, Sjoebring, U, Bjoerck, L. | | Deposit date: | 1994-08-12 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G.
Biochemistry, 33, 1994
|
|
1HRP
 
 | | CRYSTAL STRUCTURE OF HUMAN CHORIONIC GONADOTROPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN CHORIONIC GONADOTROPIN | | Authors: | Lapthorn, A.J, Harris, D.C, Isaacs, N.W. | | Deposit date: | 1994-08-15 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human chorionic gonadotropin.
Nature, 369, 1994
|
|
1HRC
 
 | |
1FRS
 
 | |
2CYK
 
 | |
1TAP
 
 | | NMR SOLUTION STRUCTURE OF RECOMBINANT TICK ANTICOAGULANT PROTEIN (RTAP), A FACTOR XA INHIBITOR FROM THE TICK ORNITHODOROS MOUBATA | | Descriptor: | FACTOR XA INHIBITOR | | Authors: | Antuch, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1994-08-16 | | Release date: | 1994-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the recombinant tick anticoagulant protein (rTAP), a factor Xa inhibitor from the tick Ornithodoros moubata.
FEBS Lett., 352, 1994
|
|
