3W4K
 
 | | Crystal Structure of human DAAO in complex with coumpound 13 | | Descriptor: | 3-hydroxy-6-(2-phenylethyl)pyridazin-4(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W5I
 
 | |
3W5X
 
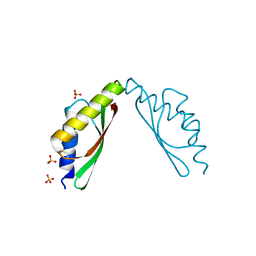 | | MamM-CTD | | Descriptor: | Magnetosome protein MamM, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2013-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain
Plos One, 9, 2014
|
|
3W64
 
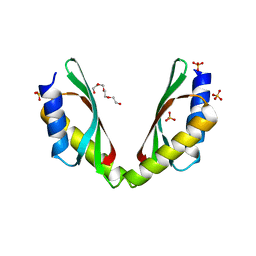 | | MamM-CTD 215-293 | | Descriptor: | Magnetosome protein MamM, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2013-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain
Plos One, 9, 2014
|
|
3W93
 
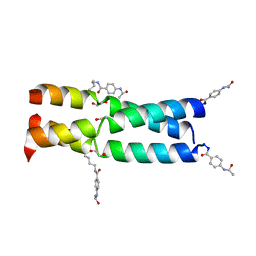 | |
3W75
 
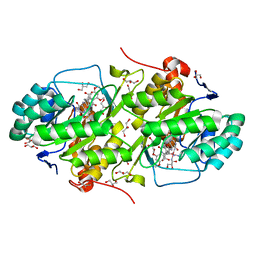 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-4-149 | | Descriptor: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-4-149
To be Published
|
|
3W7J
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-6-040 | | Descriptor: | 5-[2-(6-carboxynaphthalen-2-yl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-6-040
To be Published
|
|
3W8V
 
 | | Crystal Structure Analysis of the synthetic GCN4 coiled coil peptide | | Descriptor: | 1,2-ETHANEDIOL, GCN4n coiled coil peptide, PARA ACETAMIDO BENZOIC ACID | | Authors: | Shahar, A, Zarivach, R, Ashkenasy, G. | | Deposit date: | 2013-03-21 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-resolution structure that provides insight into coiled-coil thiodepsipeptide dynamic chemistry
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3W9I
 
 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug resistance protein MexB | | Authors: | Sakurai, K, Nakashima, R, Hayashi, K, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3W34
 
 | | Ternary complex of Thermus thermophilus HB8 uridine-cytidine kinase with substrates | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Biochemical Studies on the Reaction Mechanism of Uridine-Cytidine Kinase
Protein J., 34, 2015
|
|
3W41
 
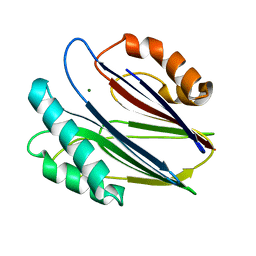 | | Crystal structure of RsbX in complex with magnesium in space group P21 | | Descriptor: | MAGNESIUM ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W2X
 
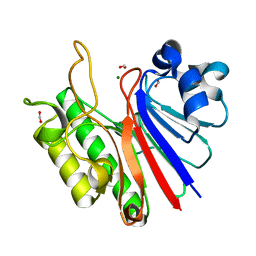 | |
3W42
 
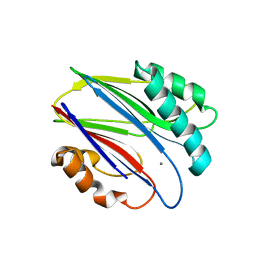 | | Crystal structure of RsbX in complex with manganese in space group P1 | | Descriptor: | MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6CX9
 
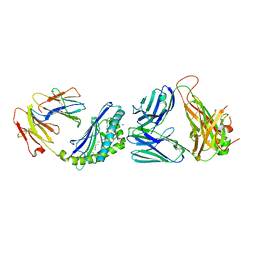 | | Structure of alpha-GSA[16,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
3W60
 
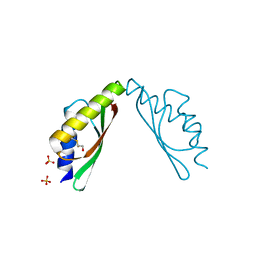 | | MamM-CTD H264A | | Descriptor: | Magnetosome protein MamM, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2013-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain
Plos One, 9, 2014
|
|
3W71
 
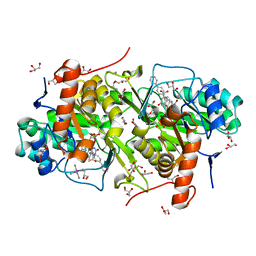 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-4-097 | | Descriptor: | 2,6-dioxo-5-[2-(4-phenylphenyl)ethyl]-1,2,3,6- tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-4-097
To be Published
|
|
3W7G
 
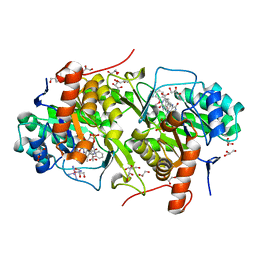 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-013 | | Descriptor: | 5-[2-(2-methoxyphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-013
To be Published
|
|
3W79
 
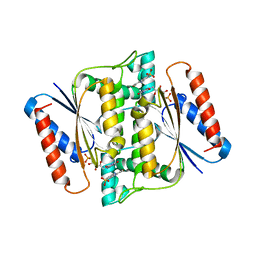 | | Crystal Structure of azoreductase AzrC in complex with sulfone-modified azo dye Orange I | | Descriptor: | 4-[(E)-(4-hydroxynaphthalen-1-yl)diazenyl]benzenesulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Ogata, D, Yu, J, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W8O
 
 | | Crystal Structure of HasAp with Iron Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, [29H,31H-phthalocyaninato(2-)-kappa~4~N~29~,N~30~,N~31~,N~32~]iron | | Authors: | Shirataki, C, Shoji, O, Terada, M, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-20 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3W7M
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with TT2-3-063 | | Descriptor: | 5-{2-[6-(methoxycarbonyl)naphthalen-2-yl]ethyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with TT2-3-063
To be Published
|
|
3W44
 
 | | Crystal structure of RsbX, selenomethionine derivative | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W8M
 
 | | Crystal Structure of HasAp with Iron Salophen | | Descriptor: | 2,2'-[1,2-PHENYLENEBIS(NITRILOMETHYLIDYNE)]BIS[PHENOLATO]](2-)-N,N',O,O']-IRON, Heme acquisition protein HasAp | | Authors: | Shirataki, C, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3W9J
 
 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug resistance protein MexB, [{2-[({[(3R)-1-{8-[(4-tert-butyl-1,3-thiazol-2-yl)carbamoyl]-4-oxo-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-4H-pyrido[1,2-a]pyrimidin-2-yl}piperidin-3-yl]oxy}carbonyl)amino]ethyl}(dimethyl)ammonio]acetate | | Authors: | Sakurai, K, Nakashima, R, Hayashi, K, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3W5Z
 
 | | MamM-CTD D249A | | Descriptor: | Magnetosome protein MamM, SODIUM ION, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2013-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain
Plos One, 9, 2014
|
|
2Q9I
 
 | |
