1XGE
 
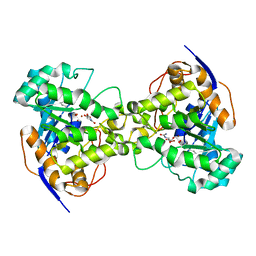 | | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between subunits | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Chan, C.W, Guss, J.M, Christopherson, R.I, Maher, M.J. | | Deposit date: | 2004-09-17 | | Release date: | 2005-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroorotase from Escherichia coli: Loop Movement and Cooperativity between Subunits
J.Mol.Biol., 348, 2005
|
|
1UBP
 
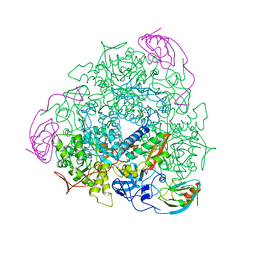 | | CRYSTAL STRUCTURE OF UREASE FROM BACILLUS PASTEURII INHIBITED WITH BETA-MERCAPTOETHANOL AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, UREASE | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1998-01-21 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The complex of Bacillus pasteurii urease with beta-mercaptoethanol from X-ray data at 1.65-A resolution
J.Biol.Inorg.Chem., 3, 1998
|
|
1S3T
 
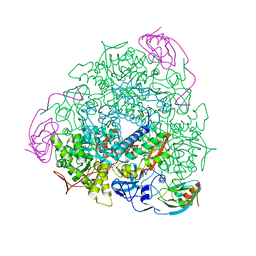 | | BORATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | BORIC ACID, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Details of Urease Inhibition by Boric Acid: Insights into the Catalytic Mechanism.
J.Am.Chem.Soc., 126, 2004
|
|
1POK
 
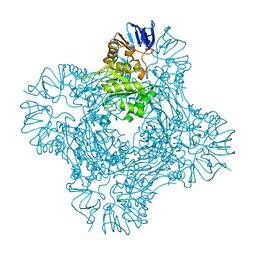 | | Crystal structure of Isoaspartyl Dipeptidase | | Descriptor: | ASPARAGINE, Isoaspartyl dipeptidase, SULFATE ION, ... | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1POJ
 
 | | Isoaspartyl Dipeptidase with bound inhibitor | | Descriptor: | 2-{[[(1S)-1-AMINO-2-CARBOXYETHYL](DIHYDROXY)PHOSPHORANYL]METHYL}-4-METHYLPENTANOIC ACID, Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1PO9
 
 | | Crytsal structure of isoaspartyl dipeptidase | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1P1M
 
 | | Structure of Thermotoga maritima amidohydrolase TM0936 bound to Ni and methionine | | Descriptor: | Hypothetical protein TM0936, METHIONINE, NICKEL (II) ION | | Authors: | Kniewel, R, Buglino, J.A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the hypothetical protein TM0936 from Thermotoga maritima at
1.5A bound to Ni and methionine
To be Published, 2003
|
|
1ONX
 
 | | Crystal structure of isoaspartyl dipeptidase from escherichia coli complexed with aspartate | | Descriptor: | ASPARTIC ACID, Isoaspartyl dipeptidase, ZINC ION | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution X-ray Structure of Isoaspartyl
Dipeptidase from Escherichia coli
Biochemistry, 42, 2003
|
|
1ONW
 
 | | Crystal structure of Isoaspartyl Dipeptidase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoaspartyl dipeptidase, ... | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution X-ray Structure of
Isoaspartyl Dipeptidase from
Escherichia coli
Biochemistry, 42, 2003
|
|
1O12
 
 | |
1NFG
 
 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
1KRC
 
 | |
1KRB
 
 | |
1KRA
 
 | |
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
1K1D
 
 | | Crystal structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Cheon, Y.H, Kim, H.S, Han, K.H, Abendroth, J, Niefind, K, Schomburg, D, Wang, J, Kim, Y. | | Deposit date: | 2001-09-25 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity.
Biochemistry, 41, 2002
|
|
1J79
 
 | | Molecular Structure of Dihydroorotase: A Paradigm for Catalysis Through the Use of a Binuclear Metal Center | | Descriptor: | N-CARBAMOYL-L-ASPARTATE, OROTIC ACID, ZINC ION, ... | | Authors: | Thoden, J.B, Phillips Jr, G.N, Neal, T.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center.
Biochemistry, 40, 2001
|
|
1J6P
 
 | |
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
1GKR
 
 | |
1GKQ
 
 | |
1GKP
 
 | | D-Hydantoinase (Dihydropyrimidinase) from Thermus sp. in space group C2221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HYDANTOINASE, SULFATE ION, ... | | Authors: | Abendroth, J, Niefind, K, Schomburg, D. | | Deposit date: | 2001-08-20 | | Release date: | 2002-06-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.295 Å) | | Cite: | X-Ray Structure of a Dihydropyrimidinase from Thermus Sp. At 1.3 A Resolution
J.Mol.Biol., 320, 2002
|
|
1FWJ
 
 | | KLEBSIELLA AEROGENES UREASE, NATIVE | | Descriptor: | NICKEL (II) ION, UREASE | | Authors: | Pearson, M.A, Karplus, P.A. | | Deposit date: | 1997-04-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Cys319 variants and acetohydroxamate-inhibited Klebsiella aerogenes urease.
Biochemistry, 36, 1997
|
|
1FWI
 
 | |
1FWH
 
 | |
