9EU7
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15b | | Descriptor: | (2-methyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EU8
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15h | | Descriptor: | (4-methyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EU9
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 15i | | Descriptor: | (4-chloranyl-1,3-thiazol-5-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EUA
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23d | | Descriptor: | (1-propylpyrazol-4-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EUB
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 24e | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [1-(2-hydroxyethyl)pyrazol-4-yl]methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EUC
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23b | | Descriptor: | (1-ethylpyrazol-4-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EUD
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23c | | Descriptor: | (1-propan-2-ylpyrazol-4-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EUE
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23a | | Descriptor: | (1-methylpyrazol-4-yl)methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9EVX
 
 | | cryoEM structure of Photosystem II averaged across S2-S3 states at 1.71 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Graca, A, Zouni, A, Messinger, J, Schroder, W.P. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (1.71 Å) | | Cite: | Cryo-electron microscopy reveals hydrogen positions and water networks in photosystem II
Science, 2024
|
|
9EY3
 
 | | The FK1 domain of FKBP51 in complex with (3S,11S,11aS)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxylic acid | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding proteins.
Chemistry, 2024
|
|
9EY4
 
 | | The FK1 domain of FKBP51 in complex with (3S,11S)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxamide | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding proteins.
Chemistry, 2024
|
|
9FA4
 
 | |
9FA7
 
 | |
9FFG
 
 | | Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Byrom, L, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography
To be published
|
|
9FFH
 
 | | Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography
To be published
|
|
9FGP
 
 | |
9FGS
 
 | | SARS-CoV-2 (wuhan variant) Spike protein in complex with the single chain fragment scFv41N (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single Chain fragment scFv41N, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of scFv41N in complex with SARS-CoV-2 Spike protein
To Be Published
|
|
9FGT
 
 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain fragment scFv76, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of scFv76 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
9FGU
 
 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76-77 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, scFv76-77 single chain fragment | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of scFv76-77 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
8Q49
 
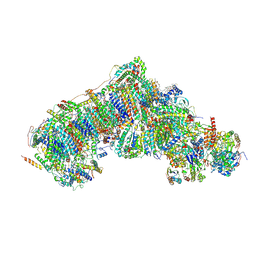 | | Outward-facing, open2 proteoliposome complex I at 2.6 A. Initially purified in LMNG. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Hirst, J. | | Deposit date: | 2023-08-05 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanism of the ischemia-induced regulatory switch in mammalian complex I
Science, 2024
|
|
8YIB
 
 | |
8V4F
 
 | |
8ZM4
 
 | |
8WMN
 
 | |
8XC1
 
 | | C. elegans SID1 in complex with dsRNA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 2024
|
|
