5H5M
 
 | | Crystal structure of HMP-1 M domain | | Descriptor: | Alpha-catenin-like protein hmp-1 | | Authors: | Kang, H, Bang, I, Weis, W.I, Choi, H.J. | | Deposit date: | 2016-11-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Caenorhabditis elegans alpha-catenin reveals constitutive binding to beta-catenin and F-actin
J. Biol. Chem., 292, 2017
|
|
1Q5A
 
 | | S-shaped trans interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
2AIN
 
 | |
2FX2
 
 | |
9ICD
 
 | | CATALYTIC MECHANISM OF NADP+-DEPENDENT ISOCITRATE DEHYDROGENASE: IMPLICATIONS FROM THE STRUCTURES OF MAGNESIUM-ISOCITRATE AND NADP+ COMPLEXES | | Descriptor: | ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hurley, J.H, Dean, A.M, Koshland Jr, D.E, Stroud, R.M. | | Deposit date: | 1991-07-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes.
Biochemistry, 30, 1991
|
|
4UT1
 
 | |
4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
4WF6
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor MK-31 | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ... | | Authors: | Maize, K.M, De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2014-09-12 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6521 Å) | | Cite: | Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates.
J.Med.Chem., 58, 2015
|
|
3EX7
 
 | |
3HHN
 
 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
7PNE
 
 | |
7PNG
 
 | |
4Q2Q
 
 | |
1FLR
 
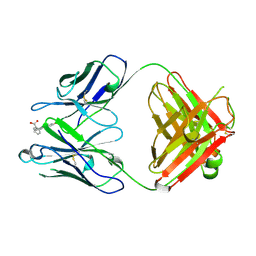 | | 4-4-20 FAB FRAGMENT | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 4-4-20 (IG*G2A=KAPPA=) FAB FRAGMENT | | Authors: | Whitlow, M. | | Deposit date: | 1995-01-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A structure of anti-fluorescein 4-4-20 Fab.
Protein Eng., 8, 1995
|
|
1FDB
 
 | |
1FDA
 
 | |
1FDD
 
 | |
1FD2
 
 | |
1GPR
 
 | |
4P0P
 
 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA, and Mg2+ | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0Q
 
 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0S
 
 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0R
 
 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA ACGTGCTTACACACAGAGGTTAGGGTGAACTT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6.501 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
5A6C
 
 | | Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation | | Descriptor: | G-PROTEIN-SIGNALING MODULATOR 2, AFADIN, SULFATE ION | | Authors: | Carminati, M, Gallini, S, Pirovano, L, Alfieri, A, Bisi, S, Mapelli, M. | | Deposit date: | 2015-06-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Concomitant Binding of Afadin to Lgn and F-Actin Directs Planar Spindle Orientation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
9F9A
 
 | | Crystal structure of MUS81-EME1 bound by compound 12. | | Descriptor: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
