2E43
 
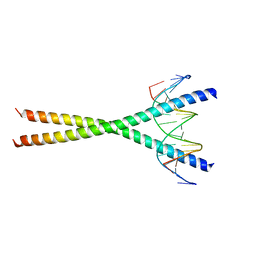 | | Crystal structure of C/EBPbeta Bzip homodimer K269A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2E42
 
 | | Crystal structure of C/EBPbeta Bzip homodimer V285A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2BM0
 
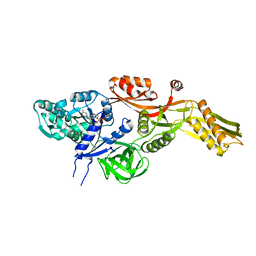 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant T84A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
2BM1
 
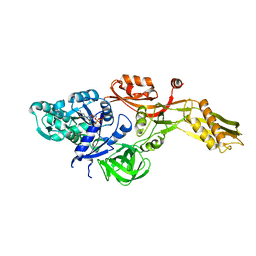 | | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant G16V | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-03-09 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Fusidic Acid Resistance and Sensitivity in EF-G
J.Mol.Biol., 348, 2005
|
|
4E75
 
 | | Structure of LpxD from Acinetobacter baumannii at 2.85A resolution (P21 form) | | Descriptor: | UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1YQM
 
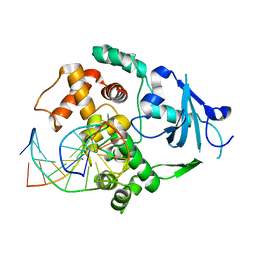 | | Catalytically inactive human 8-oxoguanine glycosylase crosslinked to 7-deazaguanine containing DNA | | Descriptor: | 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*CP*AP*(7GU)P*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
2E1P
 
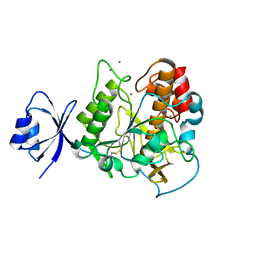 | | Crystal structure of pro-Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Saito, K, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of unautoprocessed precursor of subtilisin from a hyperthermophilic archaeon: evidence for Ca2+-induced folding
J.Biol.Chem., 282, 2007
|
|
4E19
 
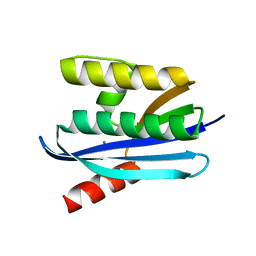 | |
1XPU
 
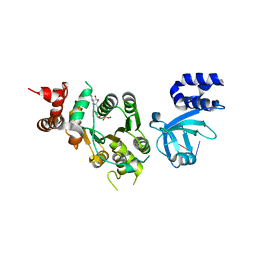 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic 5a-(3-formylphenylsulfanyl)-dihydrobicyclomycin (FPDB) | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', 5A-(3-FORMYLPHENYLSULFANYL)-DIHYDROBICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
4EB0
 
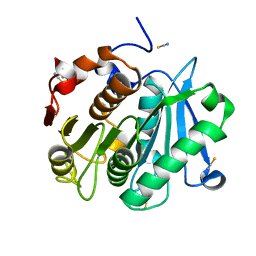 | | Crystal structure of Leaf-branch compost bacterial cutinase homolog | | Descriptor: | LCC, THIOCYANATE ION | | Authors: | Sulaiman, S, You, D.J, Eiko, K, Koga, Y, Kanaya, S. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Leaf-branch compost bacterial cutinase homolog
To be Published
|
|
1XPO
 
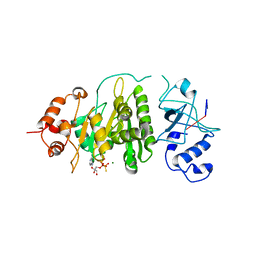 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', BICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2005-02-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
4E6U
 
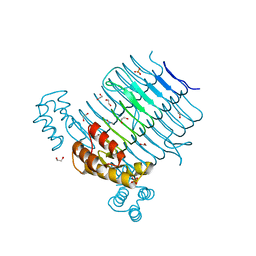 | | Structure of LpxA from Acinetobacter baumannii at 1.4A resolution (P63 form) | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-16 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4E6T
 
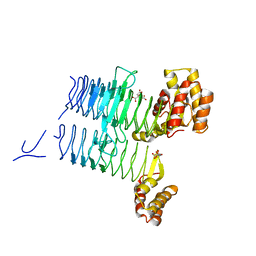 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1XVA
 
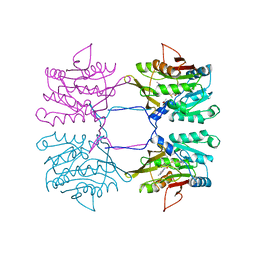 | | METHYLTRANSFERASE | | Descriptor: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
1Y0X
 
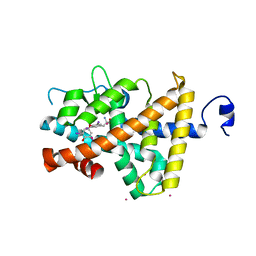 | | Thyroxine-Thyroid Hormone Receptor Interactions | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, CACODYLATE ION, Thyroid hormone receptor beta-1 | | Authors: | Sandler, B, Webb, P, Apriletti, J.W, Huber, B.R, Togashi, M, Cunha Lima, S.T, Juric, S, Nilsson, S, Wagner, R, Fletterick, R.J, Baxter, J.D. | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thyroxine-thyroid hormone receptor interactions.
J.Biol.Chem., 279, 2004
|
|
1YAO
 
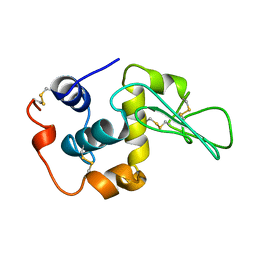 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
3QTF
 
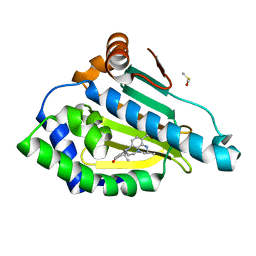 | | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity | | Descriptor: | (6S)-6,15,15,18-tetramethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-8,12-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Ingalls, C, Sutherland, A.G, Sonye, J.P, Eid, C.N, Golas, J, Liu, H, Boschelli, F, Hu, Y, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5703 Å) | | Cite: | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1XZX
 
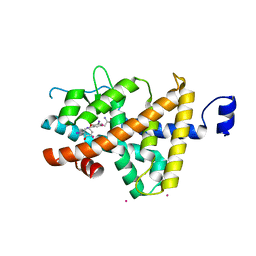 | | Thyroxine-Thyroid Hormone Receptor Interactions | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, CACODYLATE ION, Thyroid hormone receptor beta-1 | | Authors: | Sandler, B, Webb, P, Apriletti, J.W, Huber, B.R, Togashi, M, Cunha Lima, S.T, Juric, S, Nilsson, S, Wagner, R, Fletterick, R.J, Baxter, J.D. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thyroxine-thyroid hormone receptor interactions.
J.Biol.Chem., 279, 2004
|
|
1XWE
 
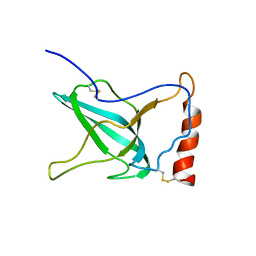 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
1YAP
 
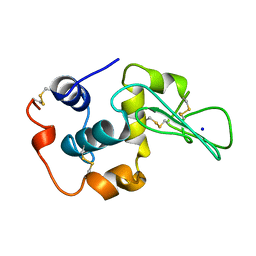 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAM
 
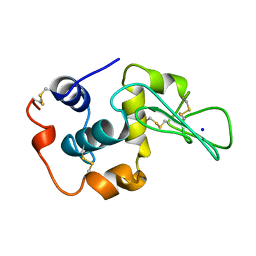 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAN
 
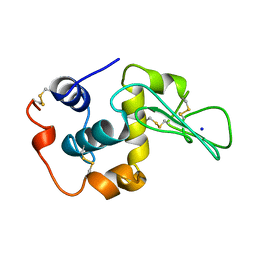 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAQ
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
4BMN
 
 | | apo structure of short-chain alcohol dehydrogenase from Ralstonia sp. DSM 6428 | | Descriptor: | 1,2-ETHANEDIOL, ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Man, H, Kulig, J, Rother, D, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
3R92
 
 | | Discovery of a macrocyclic o-aminobenzamide Hsp90 inhibitor with heterocyclic tether that shows extended biomarker activity and in vivo efficacy in a mouse xenograft model. | | Descriptor: | (3aR)-13,13,16-trimethyl-15-oxo-1,2,3,3a,4,5,12,14,15,17,18,19-dodecahydro-13H-10,6-(metheno)pyrrolo[2',1':3,4][1,4,9]triazacyclotetradecino[9,8-a]indole-7-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5801 Å) | | Cite: | Discovery of a macrocyclic o-aminobenzamide Hsp90 inhibitor with heterocyclic tether that shows extended biomarker activity and in vivo efficacy in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
