5EEK
 
 | |
7C68
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5EFZ
 
 | | Monoclinic structure of the acetyl esterase MekB | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J, Niefind, K. | | Deposit date: | 2015-10-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
7VBN
 
 | | Matrix arm of deactive state CI from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4JH7
 
 | | Crystal Structure of FosB from Bacillus cereus with Manganese and L-Cysteine-Fosfomycin Product | | Descriptor: | (2R)-2-azanyl-3-[(1R,2S)-2-oxidanyl-1-phosphono-propyl]sulfanyl-propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Thompson, M.K, Harp, J, Keithly, M.E, Jagessar, K, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-03-04 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Chemical Aspects of Resistance to the Antibiotic Fosfomycin Conferred by FosB from Bacillus cereus.
Biochemistry, 52, 2013
|
|
3NCI
 
 | | RB69 DNA Polymerase Ternary Complex with dCTP Opposite dG at 1.8 angstrom resolution | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Blaha, G, Steitz, T.A, Konigsberg, W.H, Wang, J. | | Deposit date: | 2010-06-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex.
Biochemistry, 50, 2011
|
|
3JX9
 
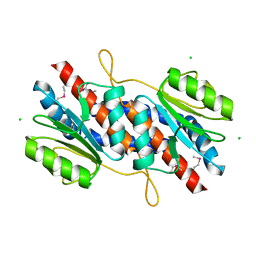 | |
5EI4
 
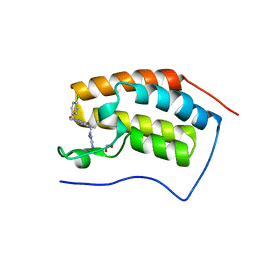 | | First domain of human bromodomain BRD4 in complex with inhibitor 8-(5-Amino-1H-[1,2,4]triazol-3-ylsulfanylmethyl)-3-(4-chlorobenzyl)-7-ethyl-3,7-dihydropurine-2,6-dione | | Descriptor: | 1,2-ETHANEDIOL, 8-[(3-azanyl-1~{H}-1,2,4-triazol-5-yl)sulfanylmethyl]-3-[(4-chlorophenyl)methyl]-7-ethyl-purine-2,6-dione, Bromodomain-containing protein 4 | | Authors: | Raux, B, Rebuffet, E, Betzi, S, Morelli, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Exploring Selective Inhibition of the First Bromodomain of the Human Bromodomain and Extra-terminal Domain (BET) Proteins.
J.Med.Chem., 59, 2016
|
|
3K0W
 
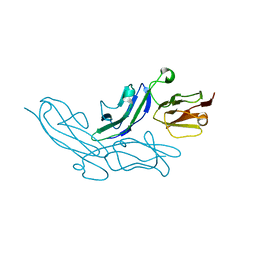 | | Crystal structure of the tandem IG-like C2-type 2 domains of the human mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Descriptor: | CHLORIDE ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, isoform 2 | | Authors: | Walker, J.R, Qiu, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tandem Ig-Like C2-Type 2 Domains of the Human Mucosa-Associated Lymphoid Tissue Lymphoma Translocation Protein 1.
To be Published
|
|
2ZKD
 
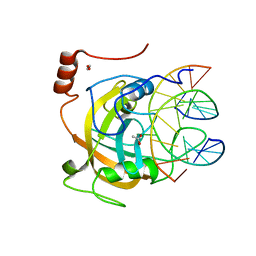 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), ... | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
7CHY
 
 | |
2WEW
 
 | | Crystal structure of human apoM in complex with myristic acid | | Descriptor: | 1,2-ETHANEDIOL, APOLIPOPROTEIN M, MYRISTIC ACID | | Authors: | Sevvana, M, Ahnstrom, J, Egerer-Sieber, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous Fatty Acid Binding Reveals the Structural Determinants for Ligand Recognition in Apolipoprotein M.
J.Mol.Biol., 393, 2009
|
|
7XJ6
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
5L5T
 
 | | Yeast 20S proteasome with human beta5i (1-138; V31M) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
8BPX
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
5EE7
 
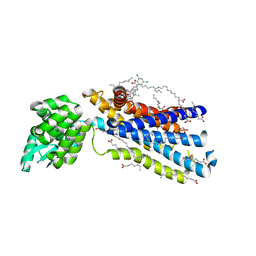 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
2ZP3
 
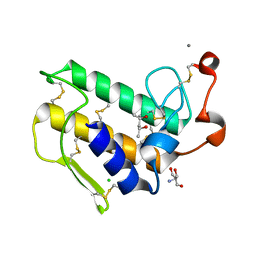 | | Carboxylic ester hydrolase, single mutant d49n of bovine pancreatic pla2 enzyme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Kanaujia, S.P, Sekar, K. | | Deposit date: | 2008-06-27 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and molecular-dynamics studies of three active-site mutants of bovine pancreatic phospholipase A(2)
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1LB0
 
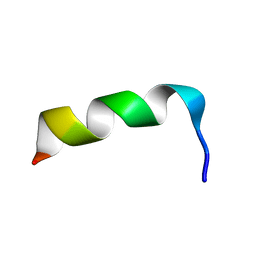 | | NMR Structure of HIV-1 gp41 659-671 13-mer peptide | | Descriptor: | GP41 | | Authors: | Biron, Z. | | Deposit date: | 2002-04-01 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Monomeric 3(10)-Helix Is Formed in Water by a 13-Residue Peptide
Representing the Neutralizing Determinant of HIV-1 on gp41(,).
Biochemistry, 41, 2002
|
|
1LQK
 
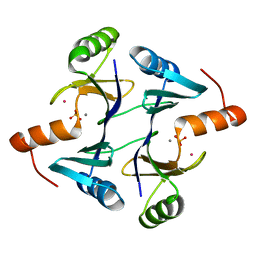 | | High Resolution Structure of Fosfomycin Resistance Protein A (FosA) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-10 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
7C6V
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to laminaritriose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
5U69
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1RKX
 
 | | Crystal Structure at 1.8 Angstrom of CDP-D-glucose 4,6-dehydratase from Yersinia pseudotuberculosis | | Descriptor: | CDP-glucose-4,6-dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Vogan, E.M, Bellamacina, C, He, X, Liu, H.W, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure at 1.8 A Resolution of CDP-d-Glucose 4,6-Dehydratase from Yersinia pseudotuberculosis
Biochemistry, 43, 2004
|
|
5L5S
 
 | |
5L6A
 
 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 17 | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
