5F9B
 
 | |
7NKE
 
 | | Crystal structure of human RXRalpha ligand binding domain in complex with 2,4-di-tert-butylphenol and a coactivator fragment | | Descriptor: | 2,4-di~{tert}-butylphenol, FORMIC ACID, Nuclear receptor coactivator 2, ... | | Authors: | Carivenc, C, Bourguet, W. | | Deposit date: | 2021-02-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2,4-Di-tert-butylphenol Induces Adipogenesis in Human Mesenchymal Stem Cells by Activating Retinoid X Receptors.
Endocrinology, 164, 2023
|
|
5VHO
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
3CCS
 
 | |
3AJH
 
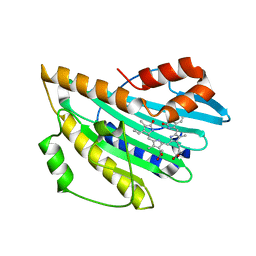 | | Crystal structure of PcyA V225D-biliverdin XIII alpha complex | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-2-ylidene]methy l]-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Wada, K, Hagiwara, Y, Fukuyama, K. | | Deposit date: | 2010-06-05 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | One residue substitution in PcyA leads to unexpected changes in tetrapyrrole substrate binding.
Biochem.Biophys.Res.Commun., 402, 2010
|
|
3ALW
 
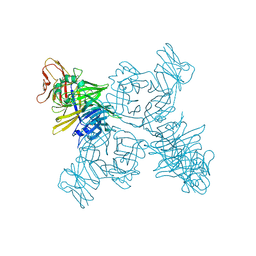 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I, MV-H-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
5H6R
 
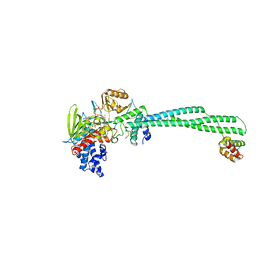 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
3H7I
 
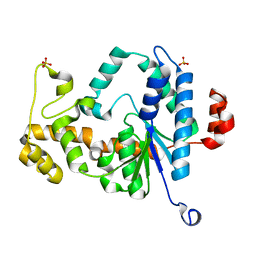 | | Structure of the metal-free D132N T4 RNase H | | Descriptor: | Ribonuclease H, SULFATE ION | | Authors: | Tomanicek, S.J, Mueser, T.C. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Additional Order Appears in the Absence of Metals in a FEN-1 protein: Structural Analysis of Magnesium Binding to Bacteriophage T4 RNaseH
To be Published
|
|
3ADE
 
 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
3CRG
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala, Glu82Asn and Lys101Ala | | Descriptor: | FORMIC ACID, GLYCEROL, Heparin-binding growth factor 1, ... | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CME
 
 | |
3H8J
 
 | |
3H8W
 
 | |
3H8S
 
 | |
3CKC
 
 | | B. thetaiotaomicron SusD | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CVI
 
 | |
2ZH6
 
 | | Complex structure of AFCCA with tRNAminiDCU and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CCA-adding enzyme, MAGNESIUM ION, ... | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2008-02-01 | | Release date: | 2008-08-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme
Embo J., 27, 2008
|
|
2Z47
 
 | |
2ZJU
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
1PE0
 
 | |
2ZM6
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kaminishi, T, Wang, H, Kawazoe, M, Ishii, R, Schluenzen, F, Hanawa-Suetsugu, K, Wilson, D.N, Nomura, M, Takemoto, C, Shirouzu, M, Fucini, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-11 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the Thermus thermophilus 30S ribosomal subunit
To be Published
|
|
3CMA
 
 | |
3CKB
 
 | | B. thetaiotaomicron SusD with maltotriose | | Descriptor: | CALCIUM ION, SusD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3CR1
 
 | | crystal structure of a minimal, mutant, all-RNA hairpin ribozyme (A38C, A-1OMA) grown from MgCl2 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3'), RNA (5'-R(*UP*CP*GP*UP*GP*GP*UP*CP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3'), ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3CQA
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala and Lys101Ala | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
