6XM8
 
 | | Crystal structure of lignostilbene bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Dioxygenase, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
7SJK
 
 | | Structure of PLS A-domain (residues 391-656) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Pls Plasmin sensitive surface protein | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-18 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SMH
 
 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
3ENS
 
 | |
6G7B
 
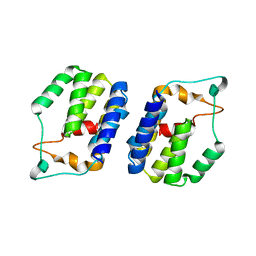 | | Nt2 domain of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | Descriptor: | ImpA-related domain protein | | Authors: | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-04-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
9CPH
 
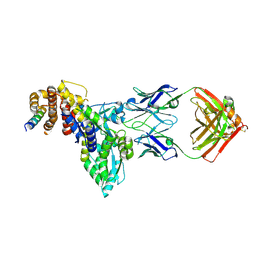 | | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | | Descriptor: | Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic antibody, ... | | Authors: | Uchikawa, E, Myasnikov, A, Dey, R, Moldoveanu, T. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis of BAK sequestration by MCL-1 in apoptosis.
Mol.Cell, 85, 2025
|
|
6N61
 
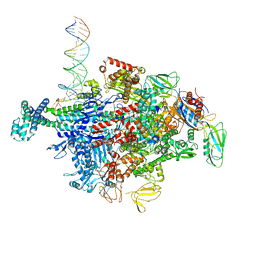 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Capistruin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capistruin, ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8HAF
 
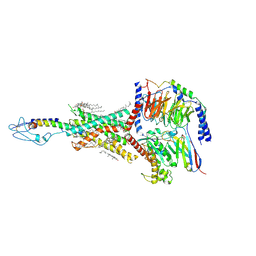 | | PTHrP-PTH1R-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
8HA0
 
 | | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
8HAO
 
 | | Human parathyroid hormone receptor-1 dimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
8BUB
 
 | | Structure of DDB1 bound to dCeMM4-engaged CDK12-cyclin K | | Descriptor: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUQ
 
 | | Structure of DDB1 bound to DS43-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[[1-(3-chlorophenyl)pyrazol-3-yl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUC
 
 | | Structure of DDB1 bound to dCeMM3-engaged CDK12-cyclin K | | Descriptor: | 2-(1~{H}-benzimidazol-2-ylsulfanyl)-~{N}-(5-chloranylpyridin-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUD
 
 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
3E8Q
 
 | | X-ray structure of rat arginase I-T135A: the unliganded complex | | Descriptor: | Arginase-1, MANGANESE (II) ION | | Authors: | Shishova, E.Y, Di Costanzo, L, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the specificity determinants of amino acid recognition by arginase.
Biochemistry, 48, 2009
|
|
8BUT
 
 | | Structure of DDB1 bound to DS61-engaged CDK12-cyclin K | | Descriptor: | 2-[[6-[[4-(2-hydroxyethyloxy)phenyl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8JN3
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 37 deg C (subparticle LLR-LRR) | | Descriptor: | Envelope protein, Human antibody DENV-115 heavy chain, Human antibody DENV-115 light chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN5
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 37 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein, Human antibody DENV-290 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN4
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-290 Fab at 4 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein, Human antibody DENV-290 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8JN2
 
 | | Cryo-EM structure of dengue virus serotype 3 strain 863DK in complex with human antibody DENV-115 Fab at 4 deg C (subparticle LLR-LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein, Human antibody DENV-115 heavy chain, ... | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
8QNI
 
 | |
6NO3
 
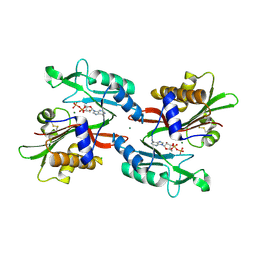 | |
6NCN
 
 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-(3-chlorophenyl)cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
8T02
 
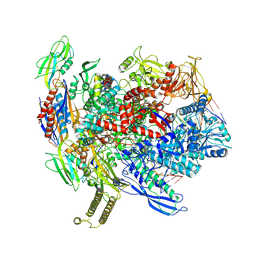 | | Reconstituted E. coli RNA polymerase post-termination complex on negatively-supercoiled DNA: unwinding duplex DNA (rPTCi) | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Brewer, J.J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-31 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | RapA opens the RNA polymerase clamp to disrupt post-termination complexes and prevent cytotoxic R-loop formation.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7B0Y
 
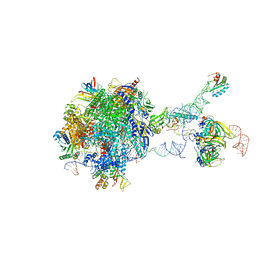 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
