9GMS
 
 | | Mtb PNPase Rv2783c | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
3AYM
 
 | |
3AYN
 
 | | Crystal structure of squid isorhodopsin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PALMITIC ACID, RETINAL, ... | | Authors: | Murakami, M, Kouyama, T. | | Deposit date: | 2011-05-09 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic Analysis of the Primary Photochemical Reaction of Squid Rhodopsin
J.Mol.Biol., 413, 2011
|
|
7TI9
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-13 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2
To Be Published
|
|
1G29
 
 | | MALK | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Diederichs, K, Diez, J, Greller, G, Mueller, C, Breed, J, Schnell, C, Vonrhein, C, Boos, W, Welte, W. | | Deposit date: | 2000-10-18 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis.
EMBO J., 19, 2000
|
|
9GMT
 
 | | Mtb PNPase Rv2783c Mutant L328F | | Descriptor: | 1-[[4-[4-[[2-phenyl-5-(trifluoromethyl)-1,3-oxazol-4-yl]carbonylamino]phenyl]phenyl]carbonylamino]cyclopentane-1-carboxylic acid, Polyribonucleotide nucleotidyltransferase, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Griesser, T, Sander, P. | | Deposit date: | 2024-08-29 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Selective inhibition of Mycobacterium tuberculosis GpsI unveils a novel strategy to target the RNA metabolism.
Nucleic Acids Res., 53, 2025
|
|
5MFQ
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-344 at 1.90 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
7V0T
 
 | | Local refinement of Band 3-I cytoplasmic domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | Band 3 anion transport protein | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V0Y
 
 | | Local refinement of Band 3-III cytoplasmic domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | Band 3 anion transport protein | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6M02
 
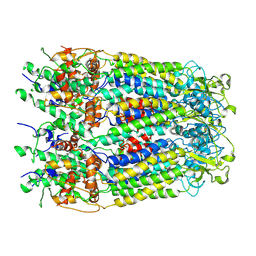 | | cryo-EM structure of human Pannexin 1 channel | | Descriptor: | Pannexin-1 | | Authors: | Ronggui, Q, Lili, D, Jilin, Z, Xuekui, Y, Lei, W, Shujia, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of human heptameric Pannexin 1 channel.
Cell Res., 30, 2020
|
|
7V0U
 
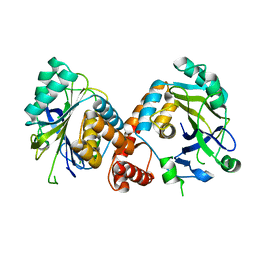 | | Local refinement of Band 3-II cytoplasmic domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | Band 3 anion transport protein | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7YWT
 
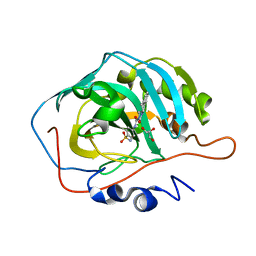 | | human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide | | Descriptor: | (11~{b}~{R})-4-oxidanylidene-~{N}-(4-sulfamoylphenyl)-3,6,7,11~{b}-tetrahydro-1~{H}-pyrazino[2,1-a]isoquinoline-2-carboxamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.106 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
2BEY
 
 | |
6JKA
 
 | | Crystal structure of metallo-beta-lactamse, IMP-1, in complex with a thiazole-bearing inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-(phenoxymethyl)-1,3-thiazol-4-yl]phenyl]propanoic acid, 6-[2-(phenoxymethyl)-1,3-thiazol-4-yl]-3,4-dihydro-1H-quinolin-2-one, Metallo-beta-lactamase type 2, ... | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
6MA3
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 2a | | Descriptor: | 4-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}butanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
7E4C
 
 | | Crystal structure of MIF bound to compound11 | | Descriptor: | 4-[(3R)-8,8-dimethyl-3,4-dihydro-2H,8H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol, CHLORIDE ION, Macrophage migration inhibitory factor, ... | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
4NYN
 
 | |
7YZH
 
 | | Schistosoma Mansoni Carbonic Anhydrase in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide, Carbonic anhydrase, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7EE8
 
 | | The crystal structure of MIF bound to compound D5 | | Descriptor: | 3-[(2R)-2-azanyl-1-oxidanyl-propyl]phenol, 3-[(2S)-2-azanyl-1-oxidanyl-propyl]phenol, CHLORIDE ION, ... | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-03-17 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Repurposing old drugs as novel inhibitors of human MIF from structural and functional analysis.
Bioorg.Med.Chem.Lett., 55, 2022
|
|
8KCP
 
 | | Cryo-EM structure of human gamma-secretase in complex with Crenigacestat | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8KCO
 
 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human gamma-secretase inhibition by anticancer clinical compounds.
Nat.Struct.Mol.Biol., 32, 2025
|
|
3BHE
 
 | | HIV-1 protease in complex with a three armed pyrrolidine derivative | | Descriptor: | N-({(3R,4R)-4-[(benzyloxy)methyl]pyrrolidin-3-yl}methyl)-N-(2-methylpropyl)benzenesulfonamide, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-11-28 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | HIV-1 protease in complex with a three armed pyrrolidine derivative
To be Published
|
|
3HMF
 
 | | Crystal Structure of the second Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein polybromo-1, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7VOB
 
 | |
5VRO
 
 | | ABA-mimicking ligand AMF1beta in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(3-fluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
