7PWJ
 
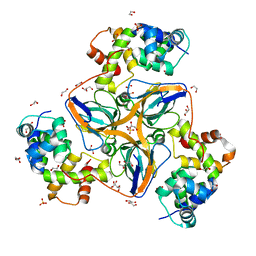 | | dUTPase from human in complex with Stl | | Descriptor: | 1,2-ETHANEDIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, ... | | Authors: | Sanz-Frasquet, C, Marina, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | The Bacteriophage-Phage-Inducible Chromosomal Island Arms Race Designs an Interkingdom Inhibitor of dUTPases.
Microbiol Spectr, 11, 2023
|
|
7PWX
 
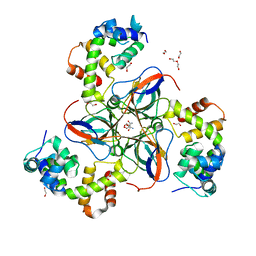 | | dUTPase from M. tuberculosis in complex with Stl | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Marina, A, Sanz-Frasquet, C. | | Deposit date: | 2021-10-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Bacteriophage-Phage-Inducible Chromosomal Island Arms Race Designs an Interkingdom Inhibitor of dUTPases.
Microbiol Spectr, 11, 2023
|
|
7BHP
 
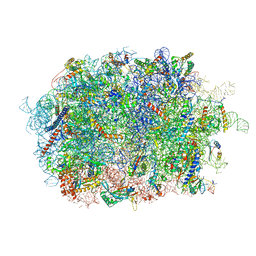 | | Cryo-EM structure of the human Ebp1 - 80S ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Desogus, J, Bhaskar, V, Chao, J.A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamic association of human Ebp1 with the ribosome.
Rna, 27, 2021
|
|
3OQG
 
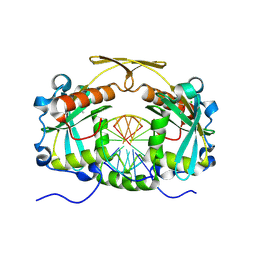 | | Restriction endonuclease HPY188I in complex with substrate DNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(*GP*AP*TP*CP*TP*GP*AP*AP*C)-3', DNA 5'-D(*GP*TP*TP*CP*AP*GP*AP*TP*C)-3', ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis.
Nucleic Acids Res., 39, 2011
|
|
5IVO
 
 | | BC2 nanobody | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BC2-nanobody | | Authors: | Braun, M.B, Stehle, T. | | Deposit date: | 2016-03-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptides in headlock - a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy.
Sci Rep, 6, 2016
|
|
3OR3
 
 | | Restriction endonuclease HPY188I in complex with product DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*T)-3', 5'-D(*GP*TP*TP*CP*A)-3', 5'-D(P*GP*AP*AP*C)-3', ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-09-06 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis.
Nucleic Acids Res., 39, 2011
|
|
2PSQ
 
 | |
2PI2
 
 | | Full-length Replication protein A subunits RPA14 and RPA32 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Borgstahl, G.E. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation.
J.Mol.Biol., 374, 2007
|
|
6KW5
 
 | | The ClassC RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome
To Be Published
|
|
8JH2
 
 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
3K8D
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
8JH3
 
 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH4
 
 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8J8D
 
 | |
6KZU
 
 | |
3K73
 
 | | Crystal Structure of Phosphate bound Holo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.5 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-10-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3K7K
 
 | | Crystal structure of the complex between Carbonic Anhydrase II and anions | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, ZINC ION, ... | | Authors: | Temperini, C. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. X-ray crystal studies of the carbonic anhydrase II-trithiocarbonate adduct-An inhibitor mimicking the sulfonamide and urea binding to the enzyme
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QKD
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
7QKC
 
 | | Crystal structure of human Cathepsin L after incubation with Sulfo-Calpeptin | | Descriptor: | Calpeptin, Cathepsin L, DI(HYDROXYETHYL)ETHER | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
2Q8S
 
 | |
2Q9C
 
 | | Structure of FTSY:GMPPNP with MGCL Complex | | Descriptor: | Cell division protein ftsY, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SULFATE ION | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
2Q9B
 
 | | Structure of FTSY:GMPPNP Complex | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein ftsY, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
2Q9Q
 
 | | The crystal structure of full length human GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Chang, Y.P, Wang, G, Chen, X.S. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of the GINS complex and functional insights into its role in DNA replication.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q9A
 
 | | Structure of Apo FTSY | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein ftsY, SULFATE ION | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
